| Full name: nitrilase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q23.3 | ||
| Entrez ID: 4817 | HGNC ID: HGNC:7828 | Ensembl Gene: ENSG00000158793 | OMIM ID: 604618 |
| Drug and gene relationship at DGIdb | |||
Expression of NIT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NIT1 | 4817 | 202891_at | 0.0736 | 0.8436 | |
| GSE20347 | NIT1 | 4817 | 202891_at | -0.1690 | 0.1040 | |
| GSE23400 | NIT1 | 4817 | 202891_at | -0.0331 | 0.5485 | |
| GSE26886 | NIT1 | 4817 | 202891_at | -0.1887 | 0.2956 | |
| GSE29001 | NIT1 | 4817 | 202891_at | 0.0301 | 0.8509 | |
| GSE38129 | NIT1 | 4817 | 202891_at | -0.0315 | 0.7973 | |
| GSE45670 | NIT1 | 4817 | 202891_at | 0.3521 | 0.0038 | |
| GSE53622 | NIT1 | 4817 | 8728 | 0.0709 | 0.2664 | |
| GSE53624 | NIT1 | 4817 | 8728 | -0.0212 | 0.7846 | |
| GSE63941 | NIT1 | 4817 | 202891_at | 1.1748 | 0.0043 | |
| GSE77861 | NIT1 | 4817 | 202891_at | 0.2266 | 0.3260 | |
| GSE97050 | NIT1 | 4817 | A_23_P376661 | -0.1374 | 0.6479 | |
| SRP007169 | NIT1 | 4817 | RNAseq | -0.0473 | 0.9028 | |
| SRP008496 | NIT1 | 4817 | RNAseq | -0.0582 | 0.8488 | |
| SRP064894 | NIT1 | 4817 | RNAseq | 0.1833 | 0.1313 | |
| SRP133303 | NIT1 | 4817 | RNAseq | 0.0862 | 0.5245 | |
| SRP159526 | NIT1 | 4817 | RNAseq | 0.2685 | 0.3842 | |
| SRP193095 | NIT1 | 4817 | RNAseq | 0.0668 | 0.4705 | |
| SRP219564 | NIT1 | 4817 | RNAseq | 0.0115 | 0.9724 | |
| TCGA | NIT1 | 4817 | RNAseq | 0.0472 | 0.4067 |
Upregulated datasets: 1; Downregulated datasets: 0.
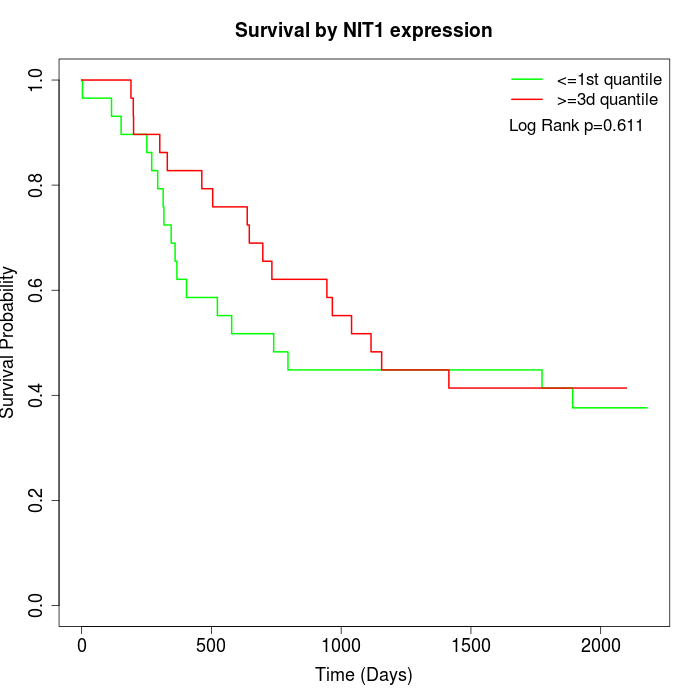
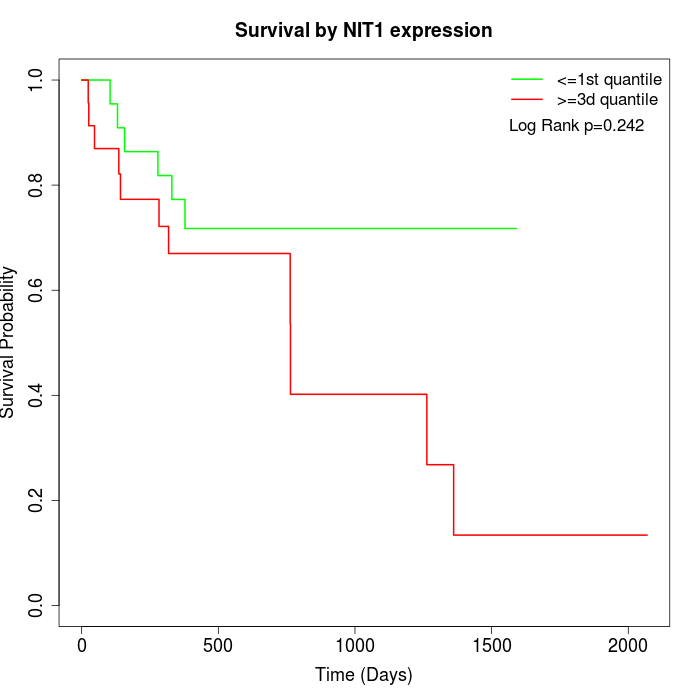
Survival by NIT1 expression:
Note: Click image to view full size file.
Copy number change of NIT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NIT1 | 4817 | 10 | 0 | 20 | |
| GSE20123 | NIT1 | 4817 | 10 | 0 | 20 | |
| GSE43470 | NIT1 | 4817 | 7 | 2 | 34 | |
| GSE46452 | NIT1 | 4817 | 2 | 1 | 56 | |
| GSE47630 | NIT1 | 4817 | 14 | 0 | 26 | |
| GSE54993 | NIT1 | 4817 | 0 | 6 | 64 | |
| GSE54994 | NIT1 | 4817 | 16 | 0 | 37 | |
| GSE60625 | NIT1 | 4817 | 0 | 0 | 11 | |
| GSE74703 | NIT1 | 4817 | 7 | 2 | 27 | |
| GSE74704 | NIT1 | 4817 | 4 | 0 | 16 | |
| TCGA | NIT1 | 4817 | 45 | 3 | 48 |
Total number of gains: 115; Total number of losses: 14; Total Number of normals: 359.
Somatic mutations of NIT1:
Generating mutation plots.
Highly correlated genes for NIT1:
Showing top 20/335 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NIT1 | INO80E | 0.781604 | 3 | 0 | 3 |
| NIT1 | MRPL38 | 0.754406 | 3 | 0 | 3 |
| NIT1 | FASTKD5 | 0.752318 | 3 | 0 | 3 |
| NIT1 | TM4SF18 | 0.749076 | 3 | 0 | 3 |
| NIT1 | NOTCH3 | 0.743691 | 3 | 0 | 3 |
| NIT1 | C8orf37 | 0.737471 | 3 | 0 | 3 |
| NIT1 | ZMYM3 | 0.731992 | 3 | 0 | 3 |
| NIT1 | RAB21 | 0.720876 | 3 | 0 | 3 |
| NIT1 | USP21 | 0.716803 | 4 | 0 | 3 |
| NIT1 | CD47 | 0.716117 | 3 | 0 | 3 |
| NIT1 | PTGES2 | 0.715991 | 3 | 0 | 3 |
| NIT1 | PHF23 | 0.707235 | 3 | 0 | 3 |
| NIT1 | SRSF7 | 0.69831 | 3 | 0 | 3 |
| NIT1 | NPRL3 | 0.697991 | 3 | 0 | 3 |
| NIT1 | CNOT1 | 0.697331 | 4 | 0 | 3 |
| NIT1 | TMEM173 | 0.694366 | 4 | 0 | 3 |
| NIT1 | ATL2 | 0.69431 | 3 | 0 | 3 |
| NIT1 | LMBR1 | 0.694298 | 3 | 0 | 3 |
| NIT1 | GIGYF1 | 0.693138 | 3 | 0 | 3 |
| NIT1 | COQ10B | 0.690472 | 3 | 0 | 3 |
For details and further investigation, click here