| Full name: neuroligin 2 | Alias Symbol: KIAA1366 | ||
| Type: protein-coding gene | Cytoband: 17p13.1 | ||
| Entrez ID: 57555 | HGNC ID: HGNC:14290 | Ensembl Gene: ENSG00000169992 | OMIM ID: 606479 |
| Drug and gene relationship at DGIdb | |||
Expression of NLGN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NLGN2 | 57555 | 226288_s_at | 0.2217 | 0.5805 | |
| GSE26886 | NLGN2 | 57555 | 226288_s_at | 0.6152 | 0.0001 | |
| GSE45670 | NLGN2 | 57555 | 226288_s_at | 0.1862 | 0.0753 | |
| GSE53622 | NLGN2 | 57555 | 46017 | 0.4046 | 0.0000 | |
| GSE53624 | NLGN2 | 57555 | 46017 | 0.4013 | 0.0000 | |
| GSE63941 | NLGN2 | 57555 | 226288_s_at | -0.2353 | 0.5014 | |
| GSE77861 | NLGN2 | 57555 | 226288_s_at | 0.0796 | 0.5987 | |
| GSE97050 | NLGN2 | 57555 | A_23_P207125 | -0.1455 | 0.5932 | |
| SRP007169 | NLGN2 | 57555 | RNAseq | 3.3205 | 0.0000 | |
| SRP008496 | NLGN2 | 57555 | RNAseq | 3.1933 | 0.0000 | |
| SRP064894 | NLGN2 | 57555 | RNAseq | 1.3587 | 0.0000 | |
| SRP133303 | NLGN2 | 57555 | RNAseq | 0.6225 | 0.0133 | |
| SRP159526 | NLGN2 | 57555 | RNAseq | 1.3246 | 0.0014 | |
| SRP193095 | NLGN2 | 57555 | RNAseq | 1.9869 | 0.0000 | |
| SRP219564 | NLGN2 | 57555 | RNAseq | 0.3733 | 0.4523 | |
| TCGA | NLGN2 | 57555 | RNAseq | 0.1659 | 0.0962 |
Upregulated datasets: 5; Downregulated datasets: 0.
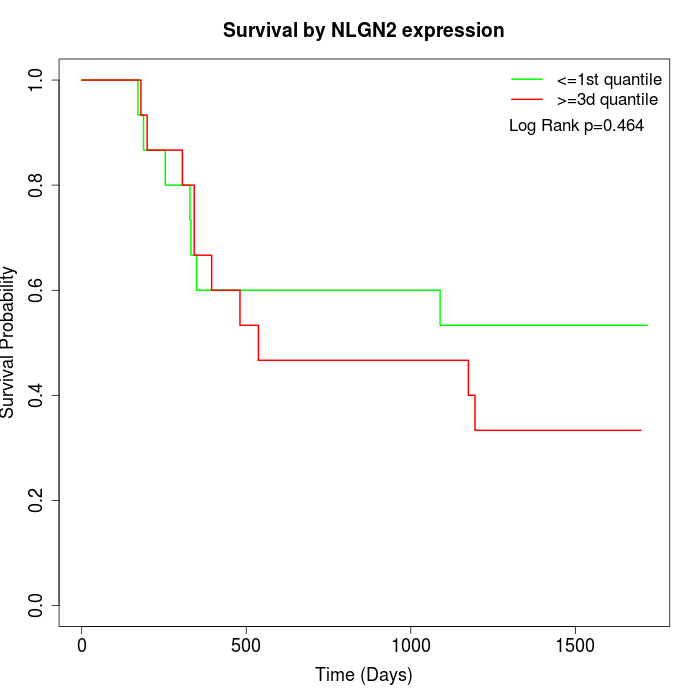
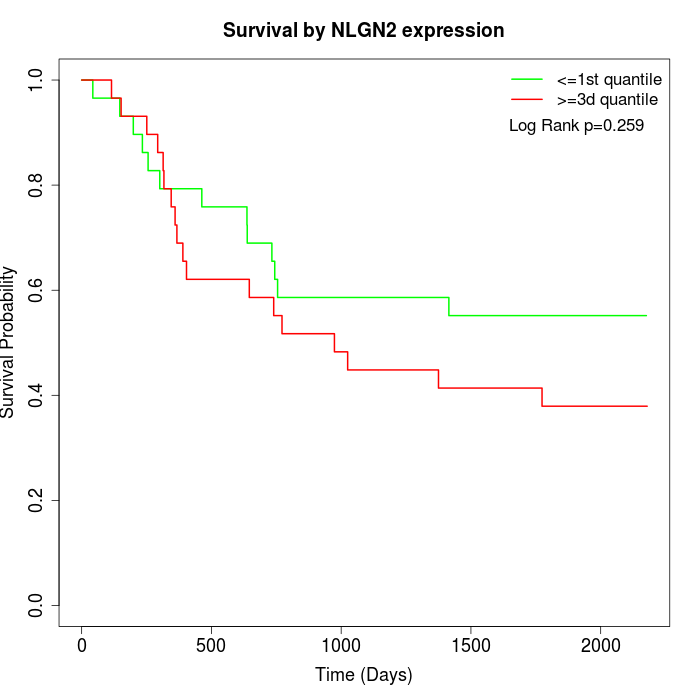
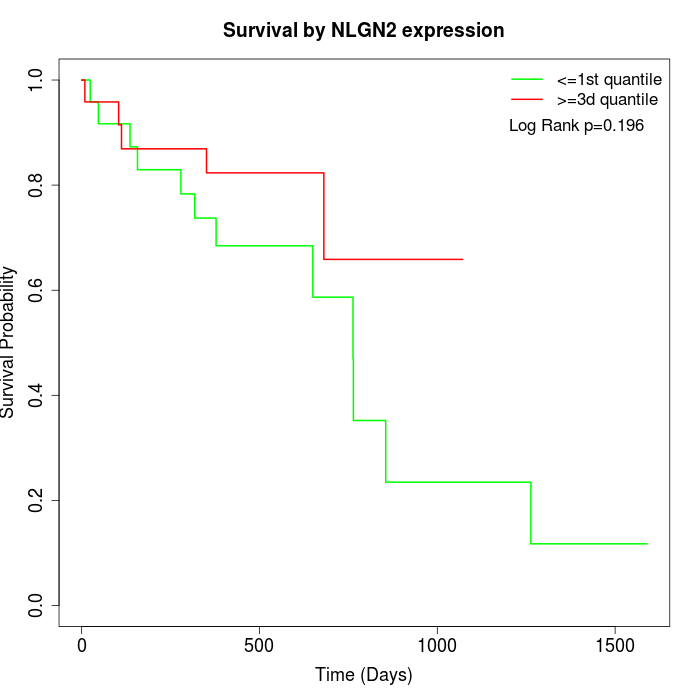
Survival by NLGN2 expression:
Note: Click image to view full size file.
Copy number change of NLGN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NLGN2 | 57555 | 4 | 2 | 24 | |
| GSE20123 | NLGN2 | 57555 | 4 | 3 | 23 | |
| GSE43470 | NLGN2 | 57555 | 1 | 4 | 38 | |
| GSE46452 | NLGN2 | 57555 | 34 | 1 | 24 | |
| GSE47630 | NLGN2 | 57555 | 7 | 1 | 32 | |
| GSE54993 | NLGN2 | 57555 | 4 | 3 | 63 | |
| GSE54994 | NLGN2 | 57555 | 5 | 8 | 40 | |
| GSE60625 | NLGN2 | 57555 | 4 | 0 | 7 | |
| GSE74703 | NLGN2 | 57555 | 1 | 2 | 33 | |
| GSE74704 | NLGN2 | 57555 | 2 | 1 | 17 | |
| TCGA | NLGN2 | 57555 | 17 | 21 | 58 |
Total number of gains: 83; Total number of losses: 46; Total Number of normals: 359.
Somatic mutations of NLGN2:
Generating mutation plots.
Highly correlated genes for NLGN2:
Showing top 20/375 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NLGN2 | DMGDH | 0.730785 | 3 | 0 | 3 |
| NLGN2 | PRKCG | 0.700337 | 4 | 0 | 4 |
| NLGN2 | MOGAT3 | 0.694653 | 4 | 0 | 4 |
| NLGN2 | OBSL1 | 0.683824 | 5 | 0 | 5 |
| NLGN2 | NKX6-3 | 0.681264 | 3 | 0 | 3 |
| NLGN2 | PCBP4 | 0.67523 | 5 | 0 | 5 |
| NLGN2 | NPB | 0.668746 | 3 | 0 | 3 |
| NLGN2 | SCUBE1 | 0.667002 | 4 | 0 | 3 |
| NLGN2 | DCDC1 | 0.663885 | 3 | 0 | 3 |
| NLGN2 | SCN1B | 0.662147 | 4 | 0 | 4 |
| NLGN2 | THBS3 | 0.661412 | 4 | 0 | 4 |
| NLGN2 | AGAP2 | 0.661411 | 3 | 0 | 3 |
| NLGN2 | NSMF | 0.658972 | 4 | 0 | 4 |
| NLGN2 | PNMA3 | 0.656789 | 3 | 0 | 3 |
| NLGN2 | PPP2R5B | 0.653726 | 6 | 0 | 5 |
| NLGN2 | OR2T1 | 0.651439 | 3 | 0 | 3 |
| NLGN2 | DSCAM | 0.649612 | 4 | 0 | 3 |
| NLGN2 | LRFN4 | 0.64781 | 3 | 0 | 3 |
| NLGN2 | FXYD2 | 0.645785 | 3 | 0 | 3 |
| NLGN2 | ISYNA1 | 0.642731 | 4 | 0 | 3 |
For details and further investigation, click here