| Full name: nicotinamide N-methyltransferase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q23.1 | ||
| Entrez ID: 4837 | HGNC ID: HGNC:7861 | Ensembl Gene: ENSG00000166741 | OMIM ID: 600008 |
| Drug and gene relationship at DGIdb | |||
Expression of NNMT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | NNMT | 4837 | 23543 | 0.4870 | 0.0011 | |
| GSE53624 | NNMT | 4837 | 23543 | 0.9334 | 0.0000 | |
| GSE97050 | NNMT | 4837 | A_23_P127584 | 0.9029 | 0.1140 | |
| SRP007169 | NNMT | 4837 | RNAseq | 3.6298 | 0.0000 | |
| SRP008496 | NNMT | 4837 | RNAseq | 4.8199 | 0.0000 | |
| SRP064894 | NNMT | 4837 | RNAseq | 0.7857 | 0.0872 | |
| SRP133303 | NNMT | 4837 | RNAseq | 1.2153 | 0.0012 | |
| SRP159526 | NNMT | 4837 | RNAseq | 0.4121 | 0.2228 | |
| SRP193095 | NNMT | 4837 | RNAseq | 2.3135 | 0.0000 | |
| SRP219564 | NNMT | 4837 | RNAseq | 1.5750 | 0.0463 | |
| TCGA | NNMT | 4837 | RNAseq | -0.0652 | 0.6412 |
Upregulated datasets: 5; Downregulated datasets: 0.
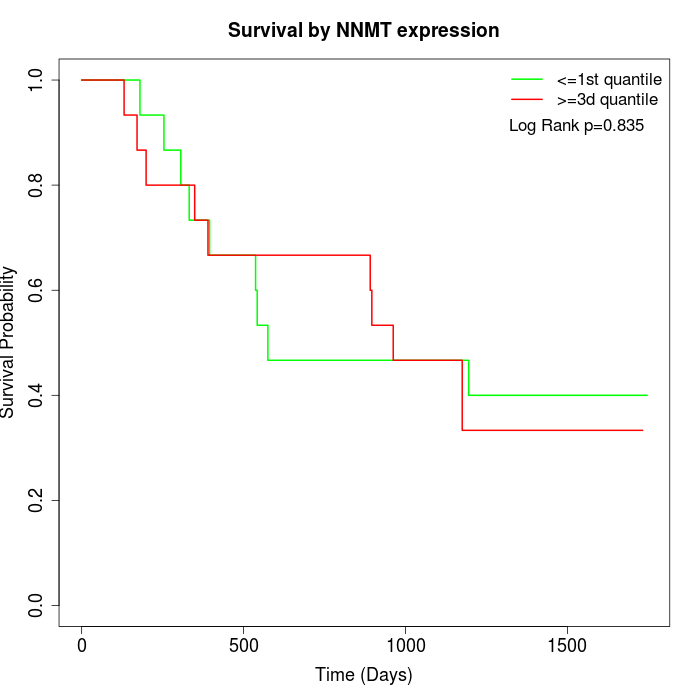
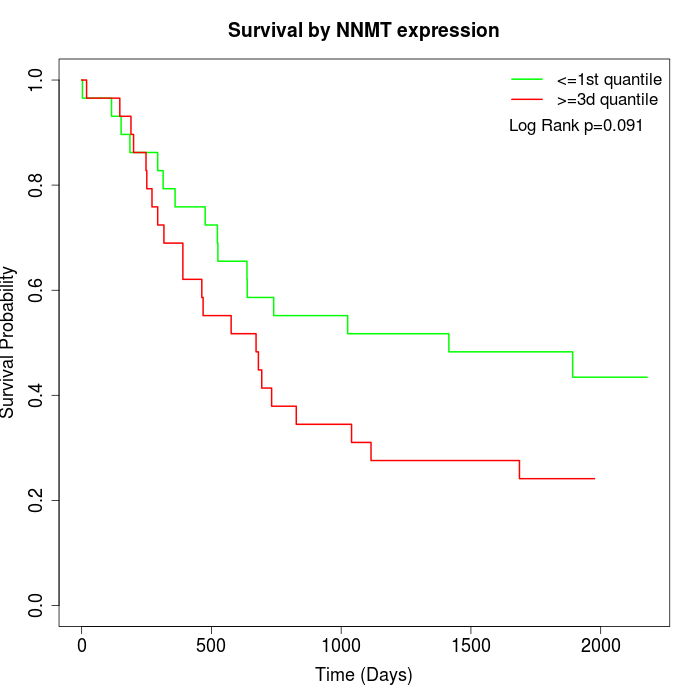
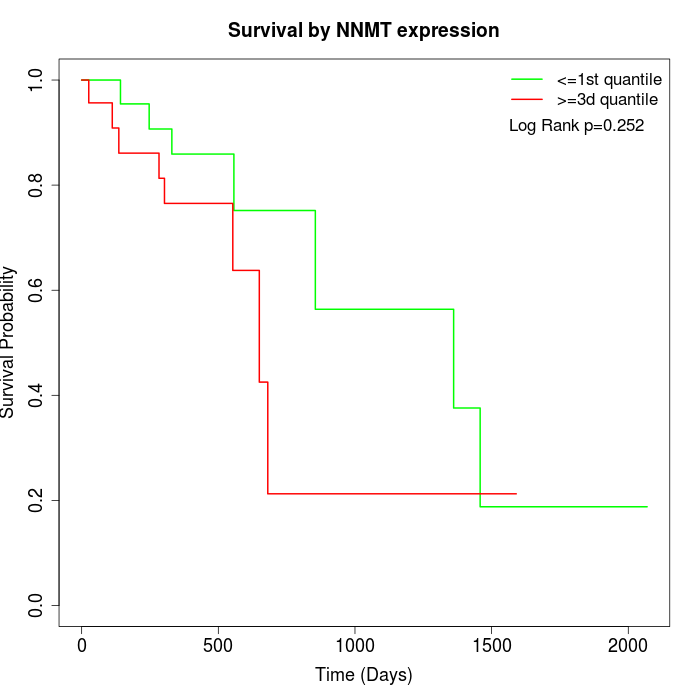
Survival by NNMT expression:
Note: Click image to view full size file.
Copy number change of NNMT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NNMT | 4837 | 0 | 13 | 17 | |
| GSE20123 | NNMT | 4837 | 0 | 13 | 17 | |
| GSE43470 | NNMT | 4837 | 2 | 7 | 34 | |
| GSE46452 | NNMT | 4837 | 3 | 26 | 30 | |
| GSE47630 | NNMT | 4837 | 2 | 19 | 19 | |
| GSE54993 | NNMT | 4837 | 10 | 0 | 60 | |
| GSE54994 | NNMT | 4837 | 5 | 19 | 29 | |
| GSE60625 | NNMT | 4837 | 0 | 3 | 8 | |
| GSE74703 | NNMT | 4837 | 1 | 5 | 30 | |
| GSE74704 | NNMT | 4837 | 0 | 9 | 11 | |
| TCGA | NNMT | 4837 | 6 | 49 | 41 |
Total number of gains: 29; Total number of losses: 163; Total Number of normals: 296.
Somatic mutations of NNMT:
Generating mutation plots.
Highly correlated genes for NNMT:
Showing top 20/274 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NNMT | DACT1 | 0.749586 | 4 | 0 | 4 |
| NNMT | DOK5 | 0.743134 | 3 | 0 | 3 |
| NNMT | TNFAIP6 | 0.74082 | 4 | 0 | 4 |
| NNMT | LHFPL2 | 0.72635 | 4 | 0 | 4 |
| NNMT | VCAN | 0.725259 | 4 | 0 | 4 |
| NNMT | TLR4 | 0.724359 | 3 | 0 | 3 |
| NNMT | FCGR3A | 0.723764 | 3 | 0 | 3 |
| NNMT | HAS2 | 0.722941 | 3 | 0 | 3 |
| NNMT | TNFRSF12A | 0.721166 | 3 | 0 | 3 |
| NNMT | THBS1 | 0.718312 | 4 | 0 | 4 |
| NNMT | PRRX1 | 0.71818 | 4 | 0 | 4 |
| NNMT | PPP1R18 | 0.71651 | 3 | 0 | 3 |
| NNMT | PLXNC1 | 0.715151 | 3 | 0 | 3 |
| NNMT | BATF3 | 0.713862 | 3 | 0 | 3 |
| NNMT | FSTL1 | 0.712884 | 4 | 0 | 4 |
| NNMT | MSC | 0.712684 | 3 | 0 | 3 |
| NNMT | PLAUR | 0.710217 | 3 | 0 | 3 |
| NNMT | TIMP1 | 0.71004 | 4 | 0 | 3 |
| NNMT | RASA3 | 0.706514 | 3 | 0 | 3 |
| NNMT | CHSY1 | 0.704905 | 4 | 0 | 3 |
For details and further investigation, click here