| Full name: neurotrimin | Alias Symbol: HNT|NTRI|IGLON2|CEPU-1 | ||
| Type: protein-coding gene | Cytoband: 11q25 | ||
| Entrez ID: 50863 | HGNC ID: HGNC:17941 | Ensembl Gene: ENSG00000182667 | OMIM ID: 607938 |
| Drug and gene relationship at DGIdb | |||
Expression of NTM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE26886 | NTM | 50863 | 241934_at | 0.0214 | 0.8850 | |
| GSE53622 | NTM | 50863 | 106132 | 0.6067 | 0.0000 | |
| GSE53624 | NTM | 50863 | 106132 | 1.0473 | 0.0000 | |
| GSE63941 | NTM | 50863 | 241934_at | -0.0083 | 0.9659 | |
| GSE97050 | NTM | 50863 | A_33_P3296333 | -0.2600 | 0.2848 | |
| SRP007169 | NTM | 50863 | RNAseq | 3.0646 | 0.0004 | |
| SRP008496 | NTM | 50863 | RNAseq | 4.0879 | 0.0000 | |
| SRP064894 | NTM | 50863 | RNAseq | 0.8783 | 0.0003 | |
| SRP133303 | NTM | 50863 | RNAseq | 1.3754 | 0.0002 | |
| SRP159526 | NTM | 50863 | RNAseq | 1.3789 | 0.0000 | |
| SRP219564 | NTM | 50863 | RNAseq | -0.0977 | 0.8992 | |
| TCGA | NTM | 50863 | RNAseq | 0.4380 | 0.0370 |
Upregulated datasets: 5; Downregulated datasets: 0.
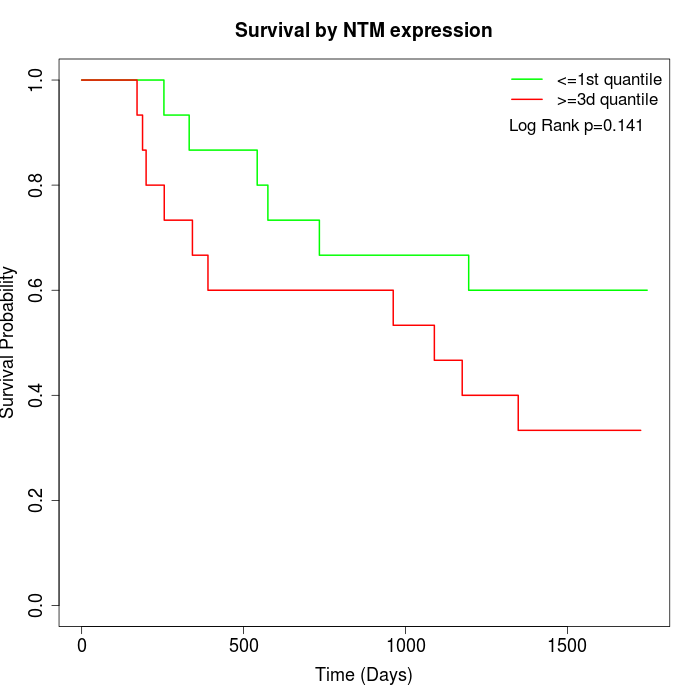
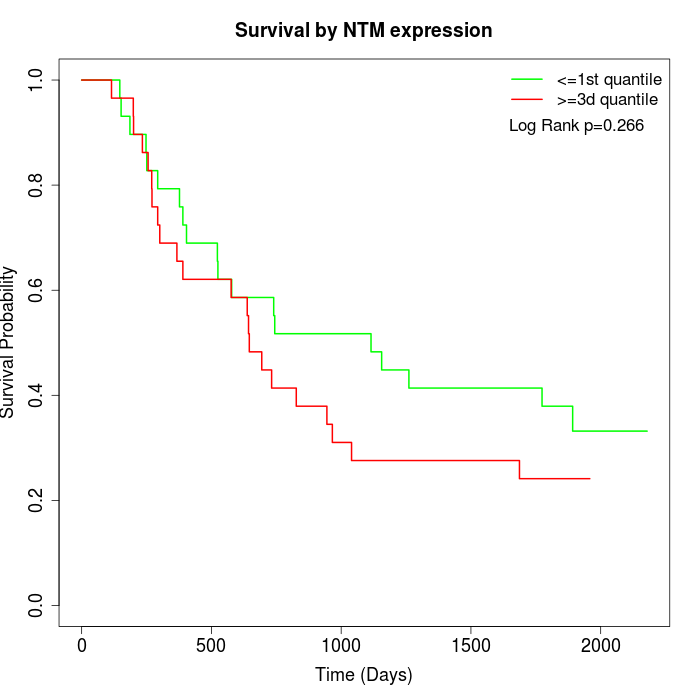
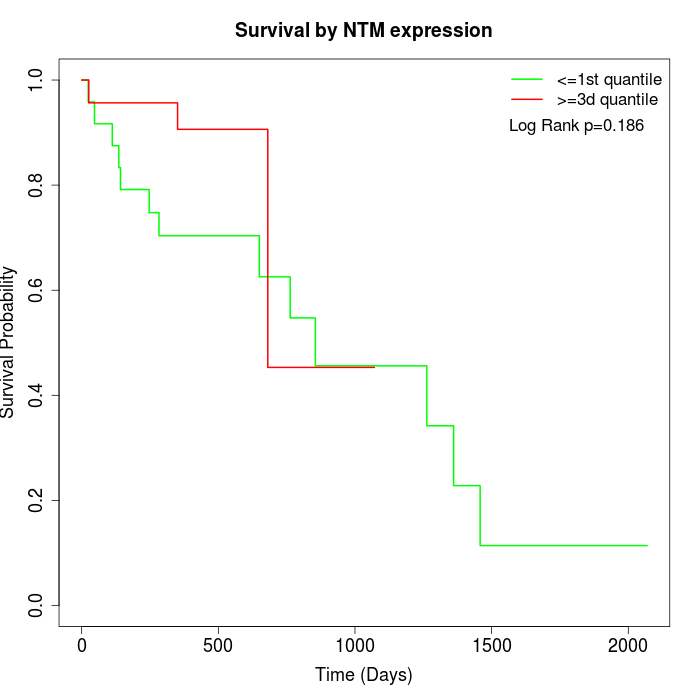
Survival by NTM expression:
Note: Click image to view full size file.
Copy number change of NTM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NTM | 50863 | 0 | 14 | 16 | |
| GSE20123 | NTM | 50863 | 0 | 14 | 16 | |
| GSE43470 | NTM | 50863 | 2 | 7 | 34 | |
| GSE46452 | NTM | 50863 | 3 | 26 | 30 | |
| GSE47630 | NTM | 50863 | 2 | 19 | 19 | |
| GSE54993 | NTM | 50863 | 10 | 0 | 60 | |
| GSE54994 | NTM | 50863 | 4 | 20 | 29 | |
| GSE60625 | NTM | 50863 | 0 | 3 | 8 | |
| GSE74703 | NTM | 50863 | 1 | 5 | 30 | |
| GSE74704 | NTM | 50863 | 0 | 10 | 10 | |
| TCGA | NTM | 50863 | 5 | 51 | 40 |
Total number of gains: 27; Total number of losses: 169; Total Number of normals: 292.
Somatic mutations of NTM:
Generating mutation plots.
Highly correlated genes for NTM:
Showing top 20/38 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NTM | ISLR | 0.739411 | 3 | 0 | 3 |
| NTM | AEBP1 | 0.721001 | 3 | 0 | 3 |
| NTM | ANTXR1 | 0.717277 | 3 | 0 | 3 |
| NTM | MXRA8 | 0.681154 | 3 | 0 | 3 |
| NTM | COL8A1 | 0.676912 | 3 | 0 | 3 |
| NTM | COL6A1 | 0.663058 | 3 | 0 | 3 |
| NTM | PDGFRL | 0.65712 | 3 | 0 | 3 |
| NTM | ITGA11 | 0.645552 | 4 | 0 | 3 |
| NTM | MICAL2 | 0.644293 | 3 | 0 | 3 |
| NTM | FSTL1 | 0.640994 | 3 | 0 | 3 |
| NTM | COL11A1 | 0.640185 | 3 | 0 | 3 |
| NTM | PLA2G12B | 0.639719 | 3 | 0 | 3 |
| NTM | RARRES2 | 0.638455 | 4 | 0 | 4 |
| NTM | SGIP1 | 0.626093 | 3 | 0 | 3 |
| NTM | LUM | 0.625083 | 3 | 0 | 3 |
| NTM | COL3A1 | 0.624743 | 3 | 0 | 3 |
| NTM | COL12A1 | 0.622939 | 3 | 0 | 3 |
| NTM | COL6A2 | 0.621153 | 3 | 0 | 3 |
| NTM | SFRP4 | 0.620418 | 3 | 0 | 3 |
| NTM | KCND2 | 0.606493 | 3 | 0 | 3 |
For details and further investigation, click here