| Full name: NUS1 dehydrodolichyl diphosphate synthase subunit | Alias Symbol: MGC7199|NgBR|TANGO14 | ||
| Type: protein-coding gene | Cytoband: 6q22.1 | ||
| Entrez ID: 116150 | HGNC ID: HGNC:21042 | Ensembl Gene: ENSG00000153989 | OMIM ID: 610463 |
| Drug and gene relationship at DGIdb | |||
Expression of NUS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NUS1 | 116150 | 225071_at | 0.3701 | 0.3629 | |
| GSE26886 | NUS1 | 116150 | 225071_at | -0.0160 | 0.9529 | |
| GSE45670 | NUS1 | 116150 | 225071_at | 0.5318 | 0.0001 | |
| GSE53622 | NUS1 | 116150 | 109523 | 0.3693 | 0.0000 | |
| GSE53624 | NUS1 | 116150 | 109523 | 0.7751 | 0.0000 | |
| GSE63941 | NUS1 | 116150 | 225071_at | -0.2749 | 0.5323 | |
| GSE77861 | NUS1 | 116150 | 225071_at | 0.4379 | 0.1219 | |
| GSE97050 | NUS1 | A_21_P0011128 | 0.1099 | 0.6706 | ||
| SRP007169 | NUS1 | 116150 | RNAseq | 0.6495 | 0.0320 | |
| SRP008496 | NUS1 | 116150 | RNAseq | 0.9322 | 0.0000 | |
| SRP064894 | NUS1 | 116150 | RNAseq | 0.4037 | 0.0398 | |
| SRP133303 | NUS1 | 116150 | RNAseq | 0.6226 | 0.0002 | |
| SRP159526 | NUS1 | 116150 | RNAseq | 0.3333 | 0.2400 | |
| SRP193095 | NUS1 | 116150 | RNAseq | 0.1052 | 0.4861 | |
| SRP219564 | NUS1 | 116150 | RNAseq | 0.4315 | 0.2089 | |
| TCGA | NUS1 | 116150 | RNAseq | -0.0489 | 0.2806 |
Upregulated datasets: 0; Downregulated datasets: 0.
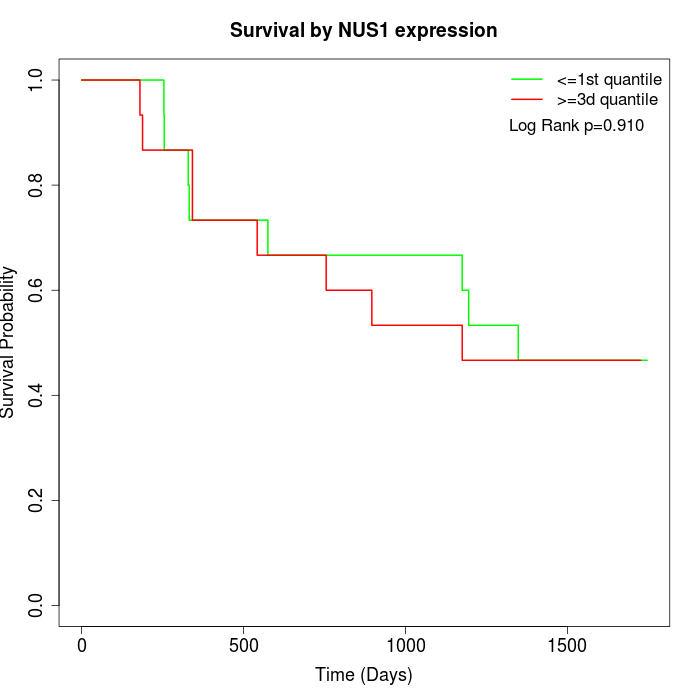
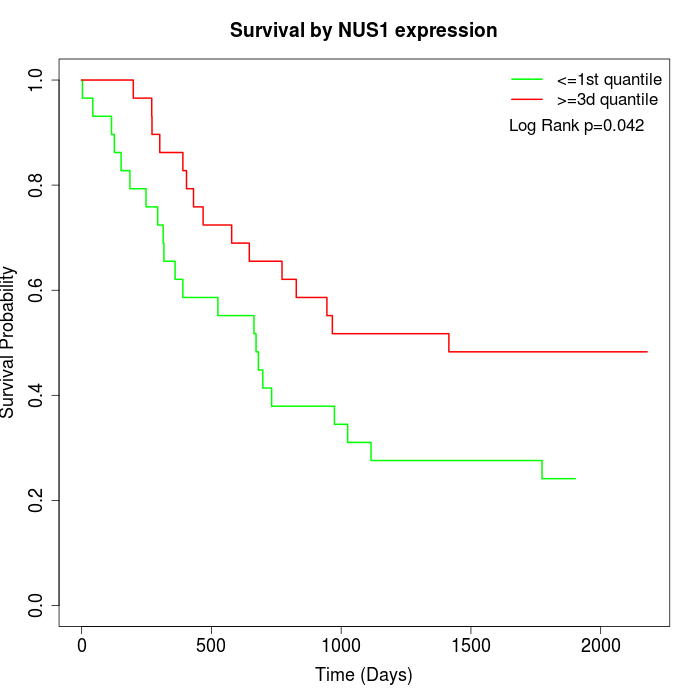
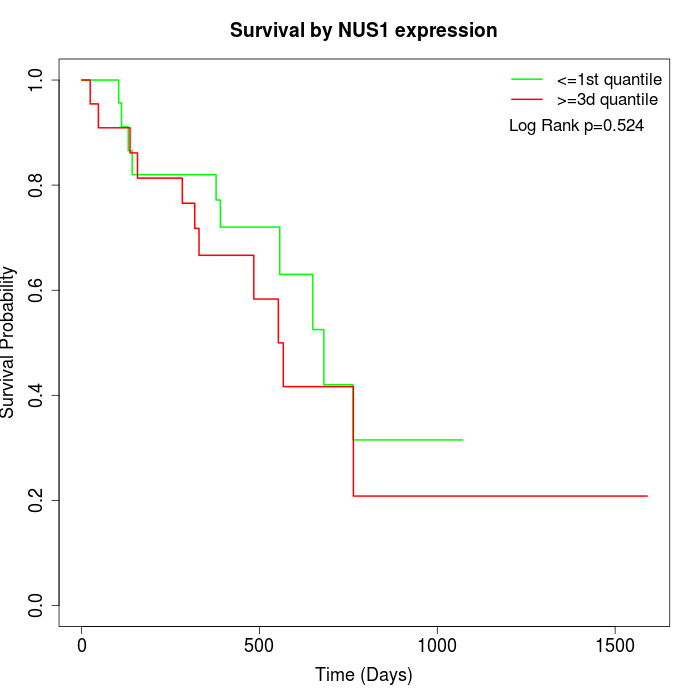
Survival by NUS1 expression:
Note: Click image to view full size file.
Copy number change of NUS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NUS1 | 116150 | 1 | 4 | 25 | |
| GSE20123 | NUS1 | 116150 | 1 | 3 | 26 | |
| GSE43470 | NUS1 | 116150 | 4 | 1 | 38 | |
| GSE46452 | NUS1 | 116150 | 2 | 11 | 46 | |
| GSE47630 | NUS1 | 116150 | 9 | 4 | 27 | |
| GSE54993 | NUS1 | 116150 | 3 | 2 | 65 | |
| GSE54994 | NUS1 | 116150 | 8 | 8 | 37 | |
| GSE60625 | NUS1 | 116150 | 0 | 1 | 10 | |
| GSE74703 | NUS1 | 116150 | 3 | 1 | 32 | |
| GSE74704 | NUS1 | 116150 | 0 | 1 | 19 | |
| TCGA | NUS1 | 116150 | 9 | 20 | 67 |
Total number of gains: 40; Total number of losses: 56; Total Number of normals: 392.
Somatic mutations of NUS1:
Generating mutation plots.
Highly correlated genes for NUS1:
Showing top 20/899 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NUS1 | BIRC3 | 0.824619 | 3 | 0 | 3 |
| NUS1 | ACP1 | 0.804346 | 4 | 0 | 4 |
| NUS1 | CTSB | 0.798548 | 3 | 0 | 3 |
| NUS1 | LINC00621 | 0.785384 | 5 | 0 | 5 |
| NUS1 | DERL1 | 0.778086 | 3 | 0 | 3 |
| NUS1 | MATR3 | 0.773687 | 3 | 0 | 3 |
| NUS1 | EIF2S1 | 0.772184 | 4 | 0 | 4 |
| NUS1 | SLC28A3 | 0.761717 | 3 | 0 | 3 |
| NUS1 | IDH3A | 0.760927 | 3 | 0 | 3 |
| NUS1 | EMG1 | 0.75974 | 3 | 0 | 3 |
| NUS1 | TMEM167A | 0.756896 | 3 | 0 | 3 |
| NUS1 | HPS5 | 0.752528 | 3 | 0 | 3 |
| NUS1 | GPD2 | 0.744353 | 3 | 0 | 3 |
| NUS1 | MSN | 0.737363 | 5 | 0 | 5 |
| NUS1 | MTX2 | 0.736265 | 3 | 0 | 3 |
| NUS1 | PSMC1 | 0.73359 | 4 | 0 | 3 |
| NUS1 | KIF2A | 0.728315 | 3 | 0 | 3 |
| NUS1 | AK2 | 0.727369 | 3 | 0 | 3 |
| NUS1 | AGTRAP | 0.725429 | 3 | 0 | 3 |
| NUS1 | NIPA2 | 0.722737 | 4 | 0 | 3 |
For details and further investigation, click here