| Full name: acid phosphatase 1 | Alias Symbol: HAAP|LMW-PTP|LMWPTP | ||
| Type: protein-coding gene | Cytoband: 2p25.3 | ||
| Entrez ID: 52 | HGNC ID: HGNC:122 | Ensembl Gene: ENSG00000143727 | OMIM ID: 171500 |
| Drug and gene relationship at DGIdb | |||
ACP1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04520 | Adherens junction |
Expression of ACP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACP1 | 52 | 215227_x_at | 0.3645 | 0.2778 | |
| GSE20347 | ACP1 | 52 | 215227_x_at | 0.6310 | 0.0040 | |
| GSE23400 | ACP1 | 52 | 215227_x_at | 0.6293 | 0.0000 | |
| GSE26886 | ACP1 | 52 | 215227_x_at | 0.2116 | 0.1195 | |
| GSE29001 | ACP1 | 52 | 215227_x_at | 0.4900 | 0.1745 | |
| GSE38129 | ACP1 | 52 | 215227_x_at | 0.8583 | 0.0000 | |
| GSE45670 | ACP1 | 52 | 215227_x_at | 0.6460 | 0.0002 | |
| GSE53622 | ACP1 | 52 | 23856 | 0.2111 | 0.0056 | |
| GSE53624 | ACP1 | 52 | 23856 | 0.2494 | 0.0003 | |
| GSE63941 | ACP1 | 52 | 215227_x_at | 0.6773 | 0.0524 | |
| GSE77861 | ACP1 | 52 | 201630_s_at | 0.2365 | 0.4758 | |
| GSE97050 | ACP1 | 52 | A_23_P56798 | -0.0178 | 0.9543 | |
| SRP007169 | ACP1 | 52 | RNAseq | 0.8854 | 0.0177 | |
| SRP008496 | ACP1 | 52 | RNAseq | 0.8744 | 0.0016 | |
| SRP064894 | ACP1 | 52 | RNAseq | 0.1974 | 0.2051 | |
| SRP133303 | ACP1 | 52 | RNAseq | 0.6399 | 0.0000 | |
| SRP159526 | ACP1 | 52 | RNAseq | 0.8099 | 0.0000 | |
| SRP193095 | ACP1 | 52 | RNAseq | 0.0802 | 0.5346 | |
| SRP219564 | ACP1 | 52 | RNAseq | 0.2907 | 0.4620 | |
| TCGA | ACP1 | 52 | RNAseq | -0.0139 | 0.7696 |
Upregulated datasets: 0; Downregulated datasets: 0.
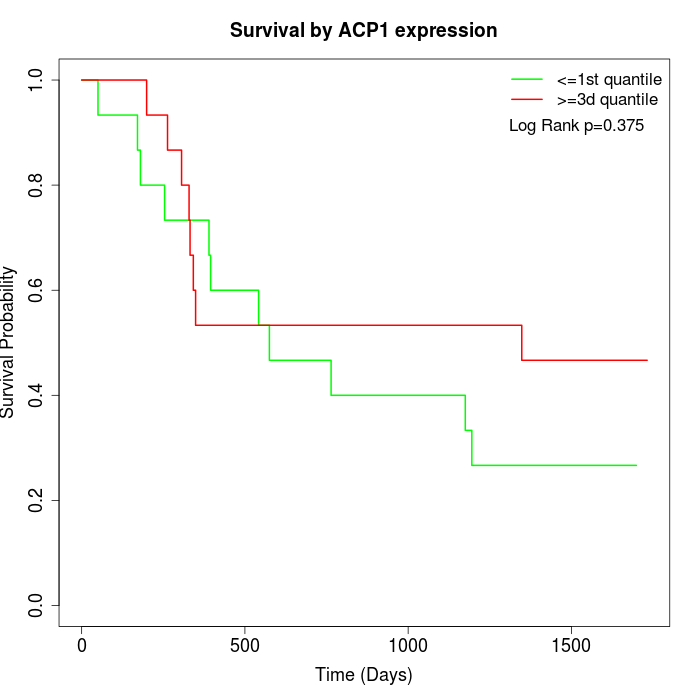
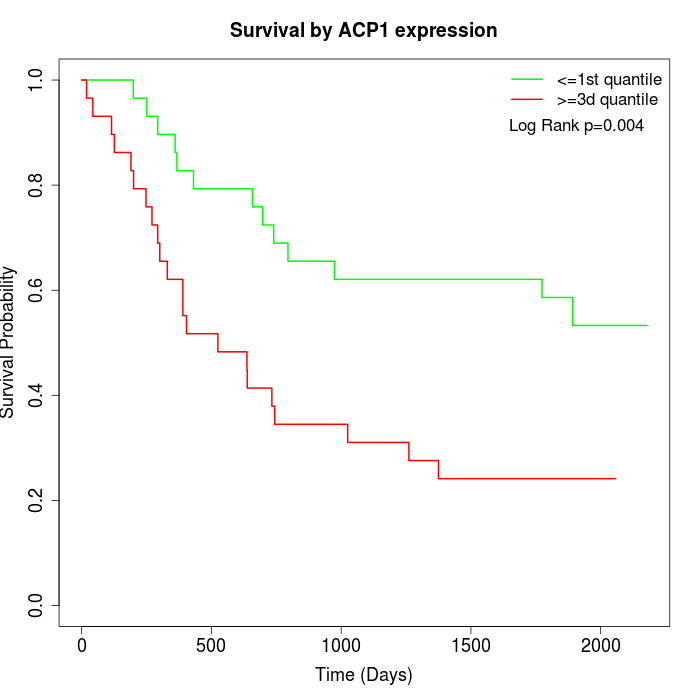
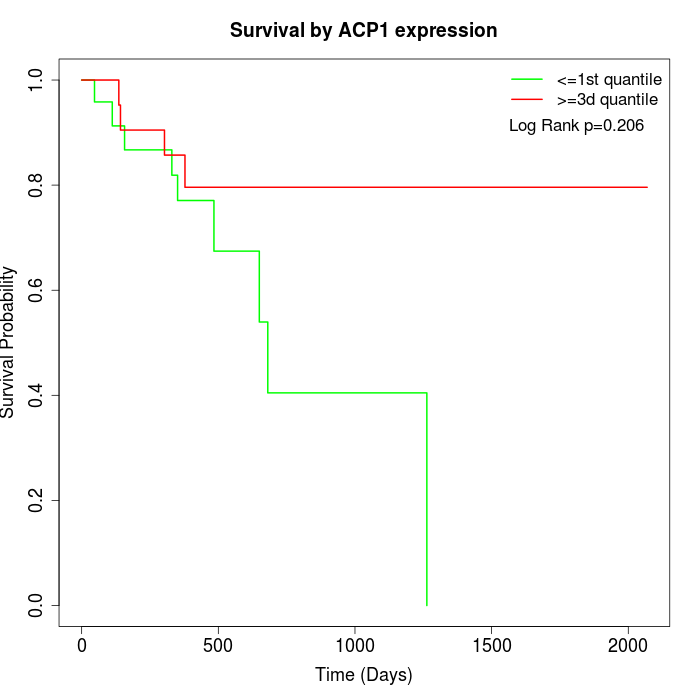
Survival by ACP1 expression:
Note: Click image to view full size file.
Copy number change of ACP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACP1 | 52 | 8 | 2 | 20 | |
| GSE20123 | ACP1 | 52 | 8 | 2 | 20 | |
| GSE43470 | ACP1 | 52 | 2 | 2 | 39 | |
| GSE46452 | ACP1 | 52 | 3 | 4 | 52 | |
| GSE47630 | ACP1 | 52 | 7 | 0 | 33 | |
| GSE54993 | ACP1 | 52 | 1 | 7 | 62 | |
| GSE54994 | ACP1 | 52 | 13 | 0 | 40 | |
| GSE60625 | ACP1 | 52 | 0 | 3 | 8 | |
| GSE74703 | ACP1 | 52 | 2 | 0 | 34 | |
| GSE74704 | ACP1 | 52 | 8 | 1 | 11 | |
| TCGA | ACP1 | 52 | 32 | 5 | 59 |
Total number of gains: 84; Total number of losses: 26; Total Number of normals: 378.
Somatic mutations of ACP1:
Generating mutation plots.
Highly correlated genes for ACP1:
Showing top 20/1394 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACP1 | NUS1 | 0.804346 | 4 | 0 | 4 |
| ACP1 | ANKS6 | 0.774511 | 3 | 0 | 3 |
| ACP1 | INTS10 | 0.758778 | 3 | 0 | 3 |
| ACP1 | ESF1 | 0.750333 | 3 | 0 | 3 |
| ACP1 | AJUBA | 0.748653 | 3 | 0 | 3 |
| ACP1 | CPSF3 | 0.740803 | 7 | 0 | 6 |
| ACP1 | ASB7 | 0.740147 | 3 | 0 | 3 |
| ACP1 | TMTC3 | 0.733996 | 3 | 0 | 3 |
| ACP1 | MASTL | 0.728821 | 5 | 0 | 5 |
| ACP1 | TUBG1 | 0.726253 | 7 | 0 | 7 |
| ACP1 | RNASEH1 | 0.722155 | 9 | 0 | 7 |
| ACP1 | IFT46 | 0.720913 | 3 | 0 | 3 |
| ACP1 | TRUB1 | 0.720357 | 4 | 0 | 4 |
| ACP1 | NEIL2 | 0.716182 | 3 | 0 | 3 |
| ACP1 | CENPBD1 | 0.71155 | 3 | 0 | 3 |
| ACP1 | NCBP1 | 0.709861 | 8 | 0 | 7 |
| ACP1 | CCT5 | 0.707554 | 8 | 0 | 7 |
| ACP1 | HNRNPC | 0.707252 | 9 | 0 | 8 |
| ACP1 | MTIF2 | 0.706472 | 8 | 0 | 7 |
| ACP1 | DDX28 | 0.700211 | 6 | 0 | 6 |
For details and further investigation, click here