| Full name: pyruvate dehydrogenase E1 alpha 1 subunit | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp22.12 | ||
| Entrez ID: 5160 | HGNC ID: HGNC:8806 | Ensembl Gene: ENSG00000131828 | OMIM ID: 300502 |
| Drug and gene relationship at DGIdb | |||
PDHA1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway |
Expression of PDHA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDHA1 | 5160 | 200980_s_at | 0.1877 | 0.4764 | |
| GSE20347 | PDHA1 | 5160 | 200980_s_at | 0.2105 | 0.1618 | |
| GSE23400 | PDHA1 | 5160 | 200980_s_at | 0.0883 | 0.3127 | |
| GSE26886 | PDHA1 | 5160 | 1555864_s_at | 0.3447 | 0.0191 | |
| GSE29001 | PDHA1 | 5160 | 200980_s_at | -0.0274 | 0.9295 | |
| GSE38129 | PDHA1 | 5160 | 200980_s_at | 0.0043 | 0.9780 | |
| GSE45670 | PDHA1 | 5160 | 200980_s_at | 0.1162 | 0.3940 | |
| GSE53622 | PDHA1 | 5160 | 18781 | -0.3546 | 0.0000 | |
| GSE53624 | PDHA1 | 5160 | 18781 | 0.1056 | 0.0562 | |
| GSE63941 | PDHA1 | 5160 | 1555864_s_at | 0.3557 | 0.5015 | |
| GSE77861 | PDHA1 | 5160 | 200980_s_at | 0.0357 | 0.8834 | |
| GSE97050 | PDHA1 | 5160 | A_23_P251095 | -0.5471 | 0.1856 | |
| SRP007169 | PDHA1 | 5160 | RNAseq | 0.2085 | 0.6343 | |
| SRP008496 | PDHA1 | 5160 | RNAseq | -0.1115 | 0.6762 | |
| SRP064894 | PDHA1 | 5160 | RNAseq | -0.0605 | 0.6201 | |
| SRP133303 | PDHA1 | 5160 | RNAseq | 0.1417 | 0.4521 | |
| SRP159526 | PDHA1 | 5160 | RNAseq | 0.1361 | 0.5512 | |
| SRP193095 | PDHA1 | 5160 | RNAseq | -0.1896 | 0.0208 | |
| SRP219564 | PDHA1 | 5160 | RNAseq | 0.0183 | 0.9673 | |
| TCGA | PDHA1 | 5160 | RNAseq | -0.2567 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
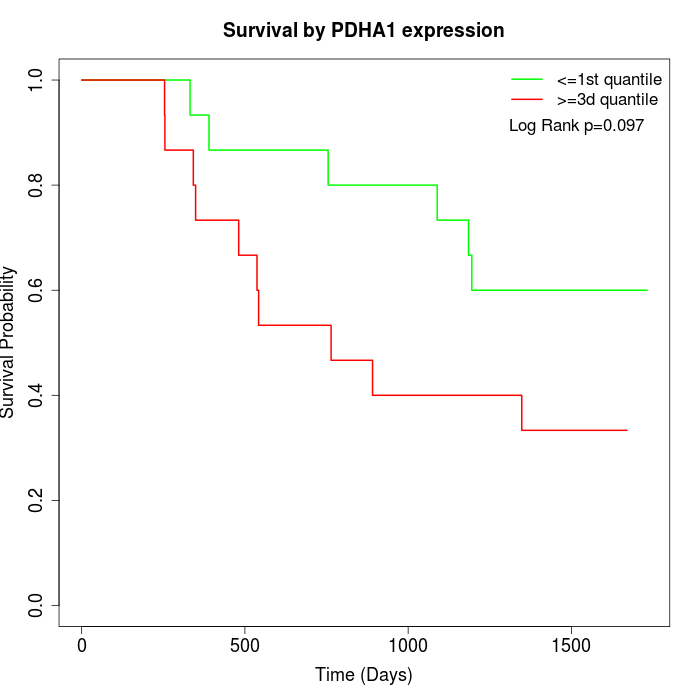
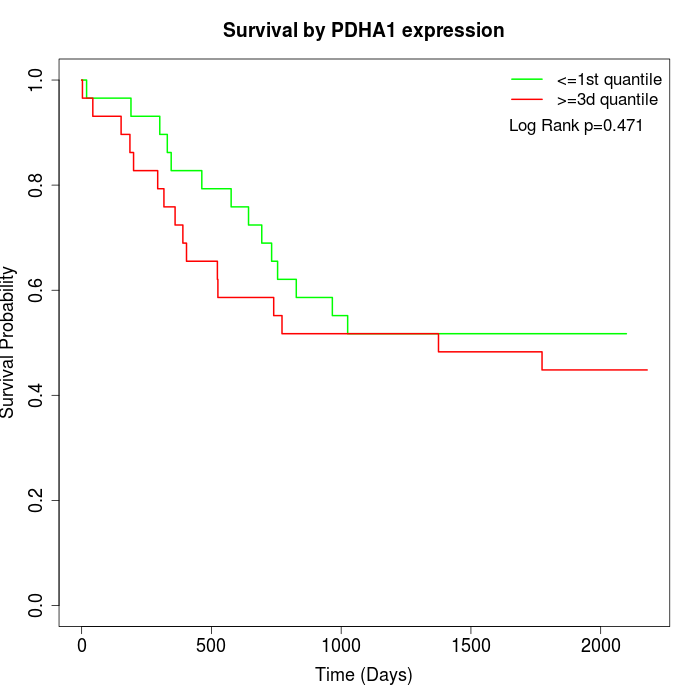
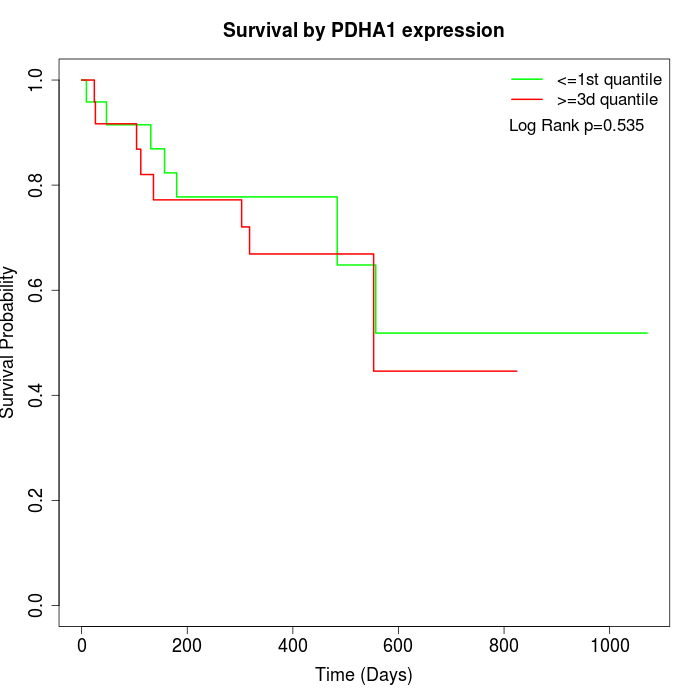
Survival by PDHA1 expression:
Note: Click image to view full size file.
Copy number change of PDHA1:
No record found for this gene.
Somatic mutations of PDHA1:
Generating mutation plots.
Highly correlated genes for PDHA1:
Showing top 20/215 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDHA1 | MLLT1 | 0.785048 | 3 | 0 | 3 |
| PDHA1 | TELO2 | 0.749604 | 3 | 0 | 3 |
| PDHA1 | SLC25A28 | 0.742482 | 3 | 0 | 3 |
| PDHA1 | SPEN | 0.732441 | 3 | 0 | 3 |
| PDHA1 | PPRC1 | 0.710518 | 3 | 0 | 3 |
| PDHA1 | MED26 | 0.707246 | 3 | 0 | 3 |
| PDHA1 | DMAP1 | 0.706804 | 3 | 0 | 3 |
| PDHA1 | TSNARE1 | 0.702413 | 4 | 0 | 3 |
| PDHA1 | ZMYND19 | 0.691883 | 3 | 0 | 3 |
| PDHA1 | NCOR2 | 0.690412 | 3 | 0 | 3 |
| PDHA1 | INO80E | 0.68865 | 3 | 0 | 3 |
| PDHA1 | DFFA | 0.687148 | 4 | 0 | 3 |
| PDHA1 | RBBP4 | 0.686609 | 3 | 0 | 3 |
| PDHA1 | NOM1 | 0.682959 | 3 | 0 | 3 |
| PDHA1 | SMARCC1 | 0.663855 | 4 | 0 | 3 |
| PDHA1 | SMNDC1 | 0.662536 | 3 | 0 | 3 |
| PDHA1 | CHMP7 | 0.662273 | 3 | 0 | 3 |
| PDHA1 | RSRC1 | 0.661018 | 3 | 0 | 3 |
| PDHA1 | MRPL43 | 0.659915 | 3 | 0 | 3 |
| PDHA1 | PIK3R3 | 0.659026 | 3 | 0 | 3 |
For details and further investigation, click here