| Full name: protein kinase N2 | Alias Symbol: PRK2|Pak-2|STK7 | ||
| Type: protein-coding gene | Cytoband: 1p22.2 | ||
| Entrez ID: 5586 | HGNC ID: HGNC:9406 | Ensembl Gene: ENSG00000065243 | OMIM ID: 602549 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PKN2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa05132 | Salmonella infection |
Expression of PKN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PKN2 | 5586 | 212628_at | -0.2881 | 0.5480 | |
| GSE20347 | PKN2 | 5586 | 212629_s_at | -0.5465 | 0.0001 | |
| GSE23400 | PKN2 | 5586 | 212628_at | -0.5544 | 0.0000 | |
| GSE26886 | PKN2 | 5586 | 212629_s_at | -1.6397 | 0.0000 | |
| GSE29001 | PKN2 | 5586 | 212629_s_at | -0.6248 | 0.0599 | |
| GSE38129 | PKN2 | 5586 | 212629_s_at | -0.4841 | 0.0061 | |
| GSE45670 | PKN2 | 5586 | 212629_s_at | -0.2780 | 0.0416 | |
| GSE53622 | PKN2 | 5586 | 18572 | -0.4830 | 0.0000 | |
| GSE53624 | PKN2 | 5586 | 18572 | -0.4524 | 0.0000 | |
| GSE63941 | PKN2 | 5586 | 212628_at | -0.1526 | 0.8282 | |
| GSE77861 | PKN2 | 5586 | 212628_at | -0.8165 | 0.0172 | |
| GSE97050 | PKN2 | 5586 | A_33_P3242833 | -0.4043 | 0.1822 | |
| SRP007169 | PKN2 | 5586 | RNAseq | -0.7826 | 0.0215 | |
| SRP008496 | PKN2 | 5586 | RNAseq | -0.7842 | 0.0000 | |
| SRP064894 | PKN2 | 5586 | RNAseq | -0.9673 | 0.0006 | |
| SRP133303 | PKN2 | 5586 | RNAseq | -0.3788 | 0.0052 | |
| SRP159526 | PKN2 | 5586 | RNAseq | -0.8055 | 0.0004 | |
| SRP193095 | PKN2 | 5586 | RNAseq | -0.6592 | 0.0000 | |
| SRP219564 | PKN2 | 5586 | RNAseq | -0.7572 | 0.0686 | |
| TCGA | PKN2 | 5586 | RNAseq | -0.0225 | 0.6268 |
Upregulated datasets: 0; Downregulated datasets: 1.
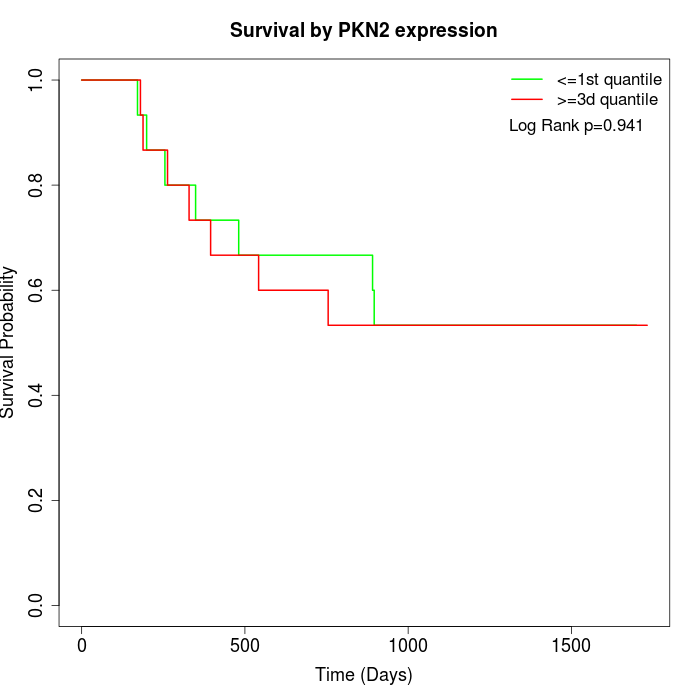
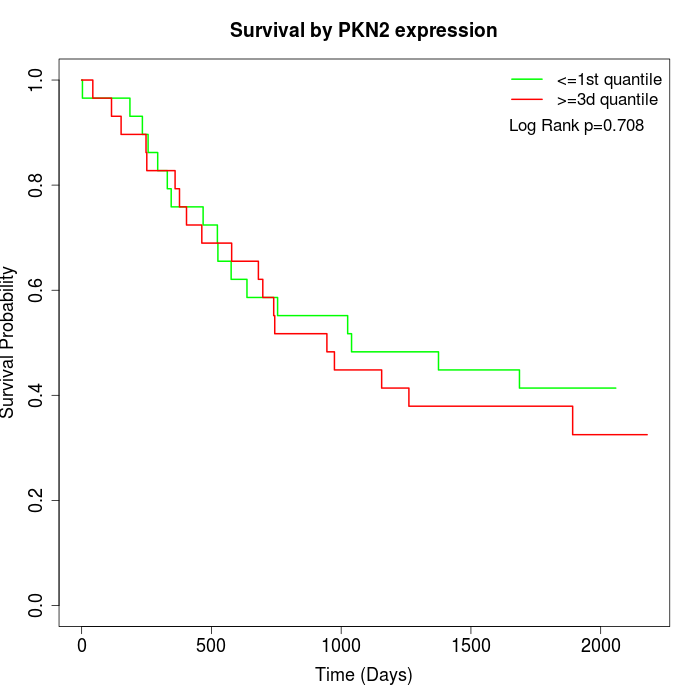
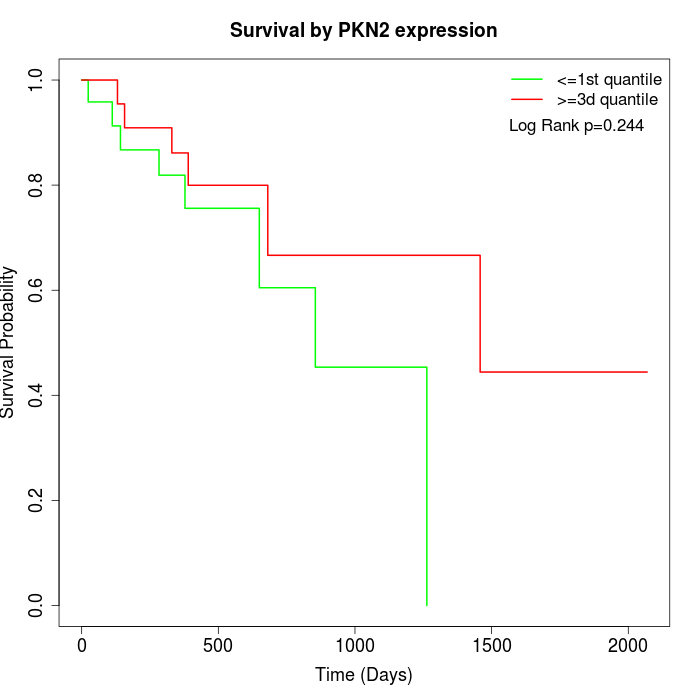
Survival by PKN2 expression:
Note: Click image to view full size file.
Copy number change of PKN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PKN2 | 5586 | 0 | 8 | 22 | |
| GSE20123 | PKN2 | 5586 | 0 | 8 | 22 | |
| GSE43470 | PKN2 | 5586 | 2 | 4 | 37 | |
| GSE46452 | PKN2 | 5586 | 1 | 1 | 57 | |
| GSE47630 | PKN2 | 5586 | 8 | 5 | 27 | |
| GSE54993 | PKN2 | 5586 | 0 | 1 | 69 | |
| GSE54994 | PKN2 | 5586 | 6 | 2 | 45 | |
| GSE60625 | PKN2 | 5586 | 0 | 0 | 11 | |
| GSE74703 | PKN2 | 5586 | 1 | 3 | 32 | |
| GSE74704 | PKN2 | 5586 | 0 | 5 | 15 | |
| TCGA | PKN2 | 5586 | 6 | 23 | 67 |
Total number of gains: 24; Total number of losses: 60; Total Number of normals: 404.
Somatic mutations of PKN2:
Generating mutation plots.
Highly correlated genes for PKN2:
Showing top 20/1383 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PKN2 | PTGR1 | 0.813787 | 3 | 0 | 3 |
| PKN2 | RBX1 | 0.795896 | 3 | 0 | 3 |
| PKN2 | IMPACT | 0.791257 | 3 | 0 | 3 |
| PKN2 | FAM122A | 0.77792 | 3 | 0 | 3 |
| PKN2 | RBM7 | 0.768954 | 3 | 0 | 3 |
| PKN2 | RBM20 | 0.763209 | 4 | 0 | 4 |
| PKN2 | FAM83D | 0.75886 | 4 | 0 | 4 |
| PKN2 | NAA20 | 0.755084 | 3 | 0 | 3 |
| PKN2 | CNOT4 | 0.751854 | 3 | 0 | 3 |
| PKN2 | SPAG17 | 0.750205 | 3 | 0 | 3 |
| PKN2 | SDHB | 0.747249 | 3 | 0 | 3 |
| PKN2 | EFHC1 | 0.743566 | 3 | 0 | 3 |
| PKN2 | ZRANB1 | 0.739809 | 3 | 0 | 3 |
| PKN2 | ZCCHC9 | 0.737719 | 4 | 0 | 4 |
| PKN2 | ALAS1 | 0.73535 | 5 | 0 | 5 |
| PKN2 | TRIM23 | 0.733202 | 4 | 0 | 4 |
| PKN2 | LYSMD3 | 0.733111 | 4 | 0 | 4 |
| PKN2 | EPG5 | 0.730942 | 5 | 0 | 4 |
| PKN2 | CCNYL1 | 0.730505 | 5 | 0 | 4 |
| PKN2 | PPIL3 | 0.728303 | 3 | 0 | 3 |
For details and further investigation, click here