| Full name: phosphoribosyl pyrophosphate amidotransferase | Alias Symbol: GPAT|PRAT | ||
| Type: protein-coding gene | Cytoband: 4q12 | ||
| Entrez ID: 5471 | HGNC ID: HGNC:9238 | Ensembl Gene: ENSG00000128059 | OMIM ID: 172450 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PPAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPAT | 5471 | 209433_s_at | 1.7423 | 0.0389 | |
| GSE20347 | PPAT | 5471 | 209433_s_at | 0.7456 | 0.1154 | |
| GSE23400 | PPAT | 5471 | 209433_s_at | 0.6806 | 0.0000 | |
| GSE26886 | PPAT | 5471 | 209433_s_at | 0.9244 | 0.0318 | |
| GSE29001 | PPAT | 5471 | 209433_s_at | 0.9139 | 0.0866 | |
| GSE38129 | PPAT | 5471 | 209433_s_at | 1.2255 | 0.0005 | |
| GSE45670 | PPAT | 5471 | 209433_s_at | 0.8832 | 0.0260 | |
| GSE53622 | PPAT | 5471 | 78261 | 0.6395 | 0.0000 | |
| GSE53624 | PPAT | 5471 | 78261 | 0.7867 | 0.0000 | |
| GSE63941 | PPAT | 5471 | 209433_s_at | 0.8407 | 0.1714 | |
| GSE77861 | PPAT | 5471 | 209433_s_at | 0.8297 | 0.0962 | |
| GSE97050 | PPAT | 5471 | A_24_P123347 | 0.4222 | 0.3046 | |
| SRP007169 | PPAT | 5471 | RNAseq | 1.3218 | 0.0152 | |
| SRP008496 | PPAT | 5471 | RNAseq | 1.1203 | 0.0001 | |
| SRP064894 | PPAT | 5471 | RNAseq | 0.4854 | 0.0813 | |
| SRP133303 | PPAT | 5471 | RNAseq | 1.1967 | 0.0000 | |
| SRP159526 | PPAT | 5471 | RNAseq | 0.7752 | 0.0398 | |
| SRP193095 | PPAT | 5471 | RNAseq | 0.5199 | 0.0003 | |
| SRP219564 | PPAT | 5471 | RNAseq | 0.5788 | 0.2222 | |
| TCGA | PPAT | 5471 | RNAseq | 0.4047 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 0.
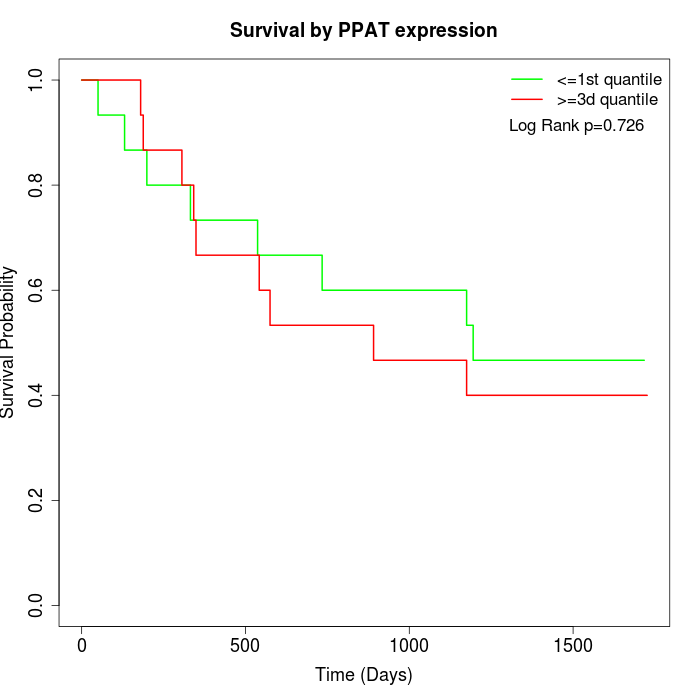
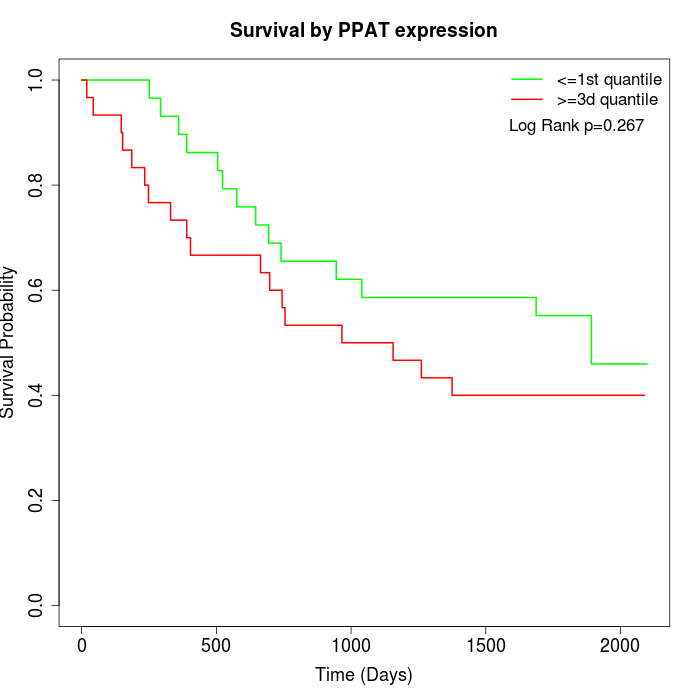
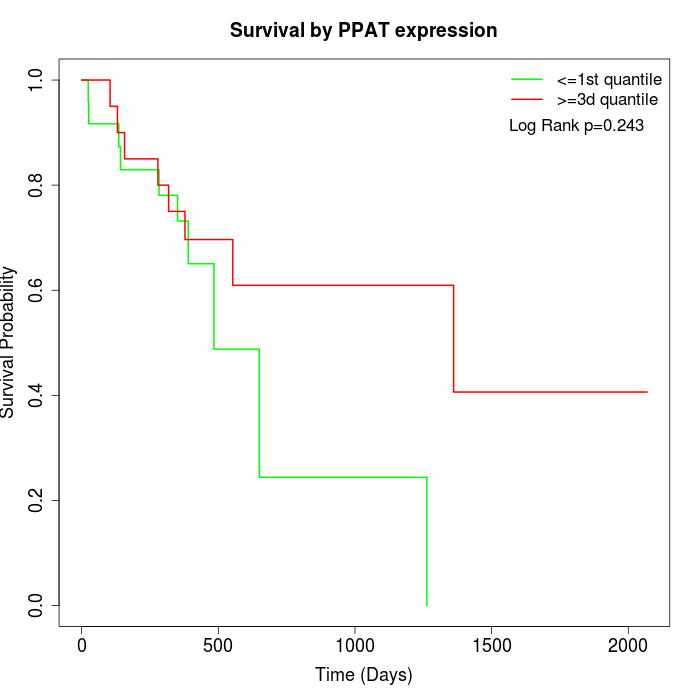
Survival by PPAT expression:
Note: Click image to view full size file.
Copy number change of PPAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPAT | 5471 | 3 | 12 | 15 | |
| GSE20123 | PPAT | 5471 | 2 | 12 | 16 | |
| GSE43470 | PPAT | 5471 | 0 | 14 | 29 | |
| GSE46452 | PPAT | 5471 | 1 | 36 | 22 | |
| GSE47630 | PPAT | 5471 | 0 | 19 | 21 | |
| GSE54993 | PPAT | 5471 | 7 | 0 | 63 | |
| GSE54994 | PPAT | 5471 | 2 | 10 | 41 | |
| GSE60625 | PPAT | 5471 | 0 | 0 | 11 | |
| GSE74703 | PPAT | 5471 | 0 | 12 | 24 | |
| GSE74704 | PPAT | 5471 | 1 | 7 | 12 | |
| TCGA | PPAT | 5471 | 20 | 27 | 49 |
Total number of gains: 36; Total number of losses: 149; Total Number of normals: 303.
Somatic mutations of PPAT:
Generating mutation plots.
Highly correlated genes for PPAT:
Showing top 20/1327 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPAT | ARHGEF19 | 0.814309 | 3 | 0 | 3 |
| PPAT | SPATA2 | 0.769451 | 3 | 0 | 3 |
| PPAT | KRTCAP2 | 0.745574 | 3 | 0 | 3 |
| PPAT | PAICS | 0.741897 | 12 | 0 | 12 |
| PPAT | SAMD1 | 0.736791 | 4 | 0 | 3 |
| PPAT | MTBP | 0.717183 | 6 | 0 | 5 |
| PPAT | TMEM181 | 0.711338 | 4 | 0 | 4 |
| PPAT | OTULIN | 0.708914 | 3 | 0 | 3 |
| PPAT | LIN9 | 0.707599 | 7 | 0 | 7 |
| PPAT | SRSF2 | 0.706726 | 3 | 0 | 3 |
| PPAT | FAM185A | 0.705545 | 3 | 0 | 3 |
| PPAT | SH2B2 | 0.702079 | 4 | 0 | 3 |
| PPAT | RAVER1 | 0.697294 | 3 | 0 | 3 |
| PPAT | ATF7IP | 0.696406 | 3 | 0 | 3 |
| PPAT | CPNE8 | 0.696335 | 4 | 0 | 3 |
| PPAT | MTRR | 0.690684 | 6 | 0 | 5 |
| PPAT | POLR1B | 0.689551 | 7 | 0 | 6 |
| PPAT | NLN | 0.686425 | 5 | 0 | 5 |
| PPAT | PTCD1 | 0.684365 | 3 | 0 | 3 |
| PPAT | DKC1 | 0.684245 | 4 | 0 | 3 |
For details and further investigation, click here