| Full name: parathymosin | Alias Symbol: ParaT | ||
| Type: protein-coding gene | Cytoband: 12p13 | ||
| Entrez ID: 5763 | HGNC ID: HGNC:9629 | Ensembl Gene: ENSG00000159335 | OMIM ID: 168440 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PTMS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PTMS | 5763 | 218045_x_at | 0.0144 | 0.9846 | |
| GSE20347 | PTMS | 5763 | 218045_x_at | 0.3766 | 0.0008 | |
| GSE23400 | PTMS | 5763 | 218045_x_at | 0.1799 | 0.0138 | |
| GSE26886 | PTMS | 5763 | 218044_x_at | 0.0952 | 0.6288 | |
| GSE29001 | PTMS | 5763 | 218045_x_at | 0.4900 | 0.1902 | |
| GSE38129 | PTMS | 5763 | 218045_x_at | 0.2401 | 0.0486 | |
| GSE45670 | PTMS | 5763 | 218044_x_at | -0.1711 | 0.0351 | |
| GSE53622 | PTMS | 5763 | 81383 | 0.1517 | 0.0129 | |
| GSE53624 | PTMS | 5763 | 81383 | 0.5048 | 0.0000 | |
| GSE63941 | PTMS | 5763 | 218045_x_at | 1.2117 | 0.0337 | |
| GSE77861 | PTMS | 5763 | 218045_x_at | 0.2549 | 0.3839 | |
| GSE97050 | PTMS | 5763 | A_33_P3333038 | -0.0832 | 0.7835 | |
| SRP007169 | PTMS | 5763 | RNAseq | 0.5679 | 0.1648 | |
| SRP008496 | PTMS | 5763 | RNAseq | 0.5282 | 0.0478 | |
| SRP064894 | PTMS | 5763 | RNAseq | 1.1320 | 0.0091 | |
| SRP133303 | PTMS | 5763 | RNAseq | 0.7031 | 0.0002 | |
| SRP159526 | PTMS | 5763 | RNAseq | 0.8395 | 0.0183 | |
| SRP193095 | PTMS | 5763 | RNAseq | 0.6906 | 0.0018 | |
| SRP219564 | PTMS | 5763 | RNAseq | 0.0492 | 0.9279 | |
| TCGA | PTMS | 5763 | RNAseq | 0.2365 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 0.
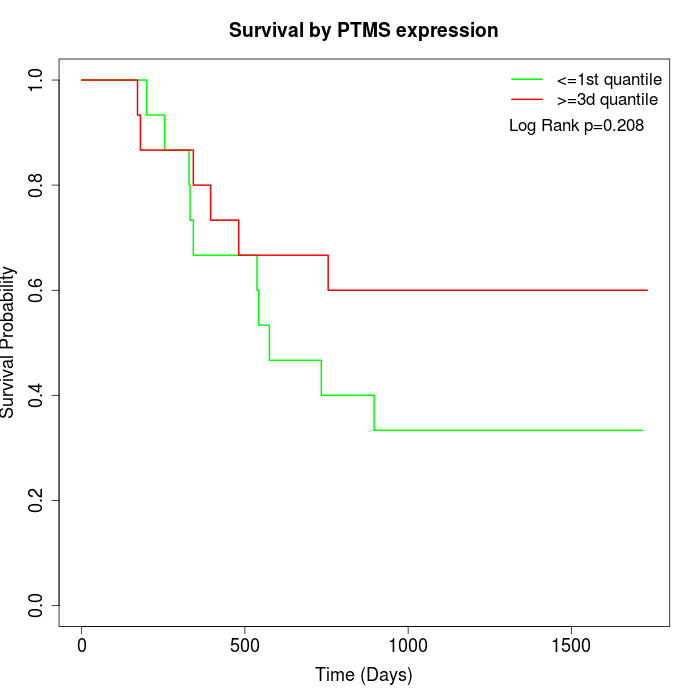
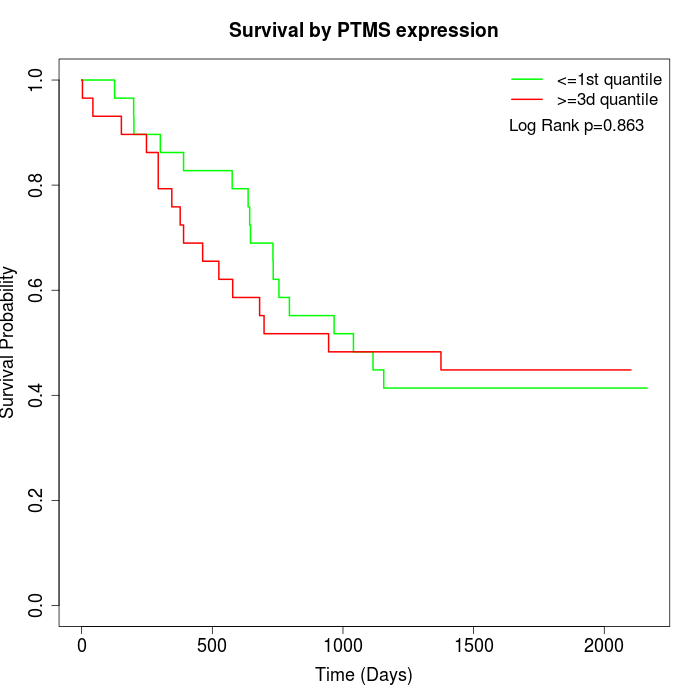
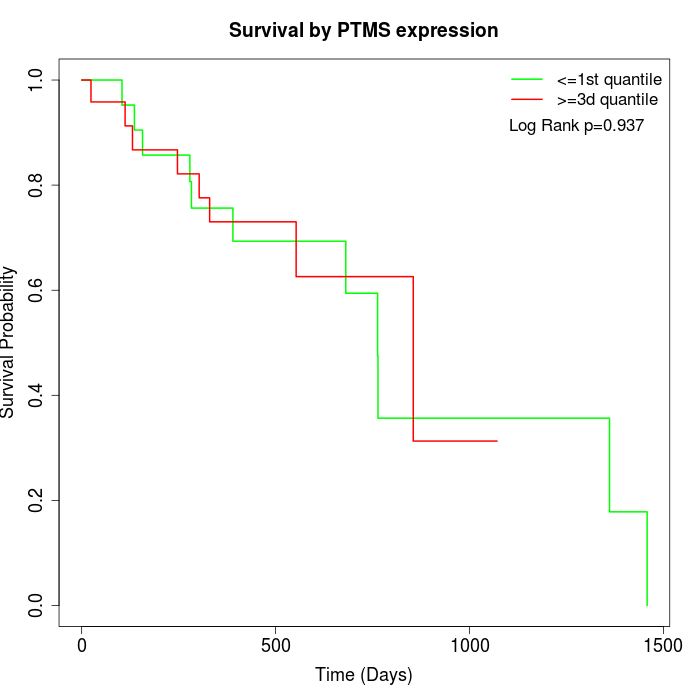
Survival by PTMS expression:
Note: Click image to view full size file.
Copy number change of PTMS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PTMS | 5763 | 9 | 3 | 18 | |
| GSE20123 | PTMS | 5763 | 9 | 3 | 18 | |
| GSE43470 | PTMS | 5763 | 8 | 3 | 32 | |
| GSE46452 | PTMS | 5763 | 10 | 1 | 48 | |
| GSE47630 | PTMS | 5763 | 12 | 2 | 26 | |
| GSE54993 | PTMS | 5763 | 1 | 10 | 59 | |
| GSE54994 | PTMS | 5763 | 10 | 2 | 41 | |
| GSE60625 | PTMS | 5763 | 0 | 1 | 10 | |
| GSE74703 | PTMS | 5763 | 7 | 2 | 27 | |
| GSE74704 | PTMS | 5763 | 5 | 2 | 13 | |
| TCGA | PTMS | 5763 | 40 | 6 | 50 |
Total number of gains: 111; Total number of losses: 35; Total Number of normals: 342.
Somatic mutations of PTMS:
Generating mutation plots.
Highly correlated genes for PTMS:
Showing top 20/244 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PTMS | RILPL1 | 0.707289 | 3 | 0 | 3 |
| PTMS | SPTAN1 | 0.691062 | 4 | 0 | 4 |
| PTMS | APC2 | 0.668839 | 5 | 0 | 4 |
| PTMS | AGAP2-AS1 | 0.665956 | 3 | 0 | 3 |
| PTMS | ATP2B4 | 0.646079 | 4 | 0 | 3 |
| PTMS | TMEM173 | 0.636768 | 3 | 0 | 3 |
| PTMS | FIBCD1 | 0.634806 | 4 | 0 | 4 |
| PTMS | ERC1 | 0.634566 | 4 | 0 | 3 |
| PTMS | SERTAD3 | 0.629332 | 3 | 0 | 3 |
| PTMS | APLP2 | 0.627112 | 5 | 0 | 3 |
| PTMS | ELMOD3 | 0.626068 | 5 | 0 | 4 |
| PTMS | TET1 | 0.625131 | 4 | 0 | 3 |
| PTMS | FPGS | 0.623568 | 3 | 0 | 3 |
| PTMS | UNC5A | 0.623328 | 4 | 0 | 3 |
| PTMS | CRMP1 | 0.620889 | 5 | 0 | 5 |
| PTMS | SF3B4 | 0.61346 | 5 | 0 | 4 |
| PTMS | SHISA4 | 0.612758 | 3 | 0 | 3 |
| PTMS | MLF2 | 0.610892 | 9 | 0 | 5 |
| PTMS | TTBK2 | 0.603785 | 5 | 0 | 4 |
| PTMS | ABL1 | 0.603636 | 6 | 0 | 4 |
For details and further investigation, click here