| Full name: ATPase plasma membrane Ca2+ transporting 4 | Alias Symbol: PMCA4 | ||
| Type: protein-coding gene | Cytoband: 1q32.1 | ||
| Entrez ID: 493 | HGNC ID: HGNC:817 | Ensembl Gene: ENSG00000058668 | OMIM ID: 108732 |
| Drug and gene relationship at DGIdb | |||
ATP2B4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa04024 | cAMP signaling pathway | |
| hsa04261 | Adrenergic signaling in cardiomyocytes |
Expression of ATP2B4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP2B4 | 493 | 212135_s_at | -0.1527 | 0.8759 | |
| GSE20347 | ATP2B4 | 493 | 212136_at | -0.0344 | 0.8616 | |
| GSE23400 | ATP2B4 | 493 | 212135_s_at | -0.0083 | 0.9592 | |
| GSE26886 | ATP2B4 | 493 | 212136_at | -0.2902 | 0.4311 | |
| GSE29001 | ATP2B4 | 493 | 212136_at | 0.3100 | 0.4105 | |
| GSE38129 | ATP2B4 | 493 | 212136_at | -0.4297 | 0.0199 | |
| GSE45670 | ATP2B4 | 493 | 212136_at | 0.0413 | 0.8629 | |
| GSE53622 | ATP2B4 | 493 | 101698 | -0.0945 | 0.4656 | |
| GSE53624 | ATP2B4 | 493 | 121264 | 0.0504 | 0.4831 | |
| GSE63941 | ATP2B4 | 493 | 212135_s_at | -0.5293 | 0.4408 | |
| GSE77861 | ATP2B4 | 493 | 212136_at | 0.9979 | 0.0013 | |
| GSE97050 | ATP2B4 | 493 | A_23_P11841 | -0.7600 | 0.2819 | |
| SRP007169 | ATP2B4 | 493 | RNAseq | 1.3932 | 0.0005 | |
| SRP008496 | ATP2B4 | 493 | RNAseq | 1.1821 | 0.0000 | |
| SRP064894 | ATP2B4 | 493 | RNAseq | 0.2717 | 0.2736 | |
| SRP133303 | ATP2B4 | 493 | RNAseq | 0.4428 | 0.1021 | |
| SRP159526 | ATP2B4 | 493 | RNAseq | 0.1306 | 0.6752 | |
| SRP193095 | ATP2B4 | 493 | RNAseq | 0.7550 | 0.0000 | |
| SRP219564 | ATP2B4 | 493 | RNAseq | -0.0914 | 0.8952 | |
| TCGA | ATP2B4 | 493 | RNAseq | -0.0367 | 0.6075 |
Upregulated datasets: 2; Downregulated datasets: 0.
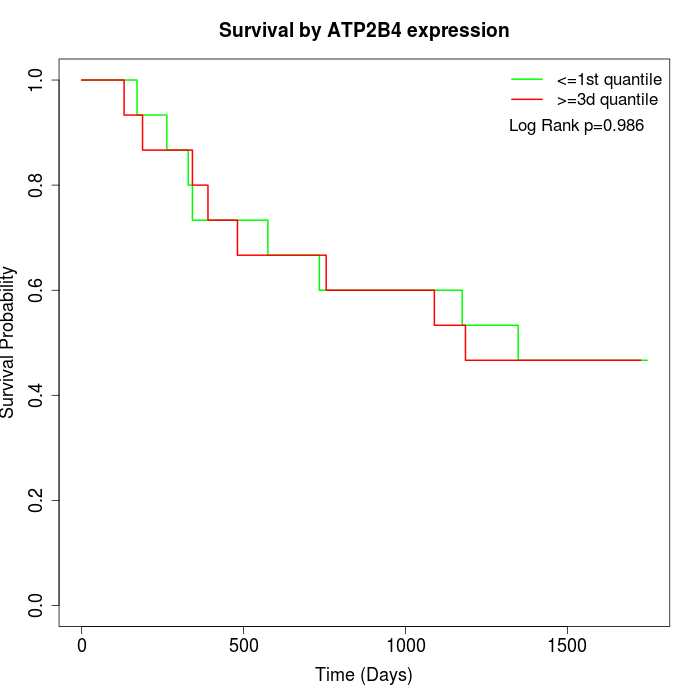
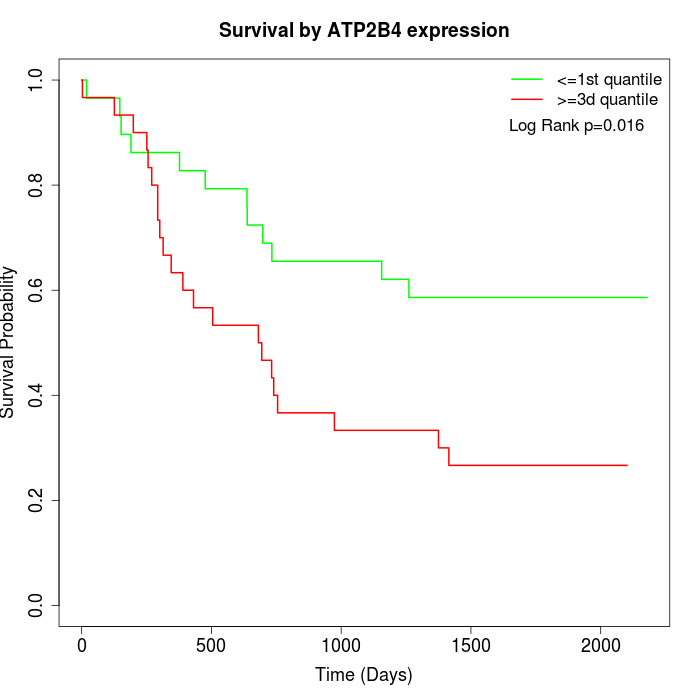
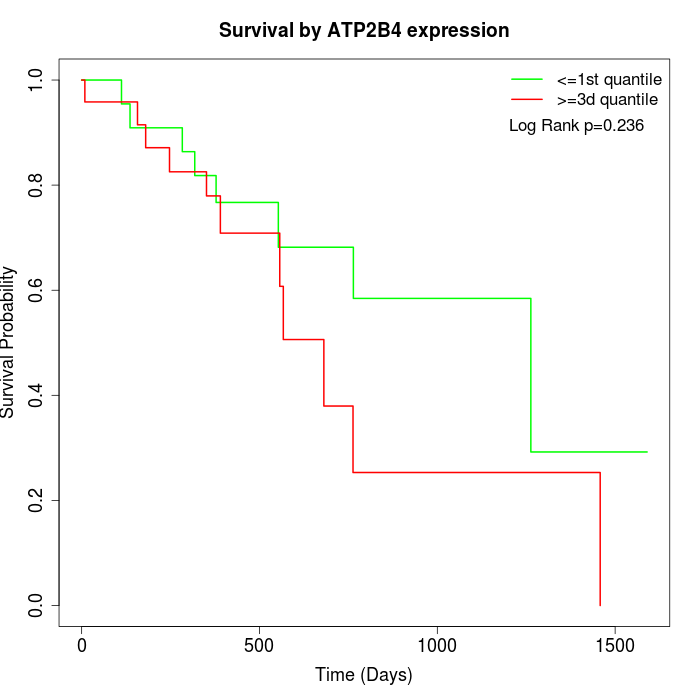
Survival by ATP2B4 expression:
Note: Click image to view full size file.
Copy number change of ATP2B4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP2B4 | 493 | 11 | 0 | 19 | |
| GSE20123 | ATP2B4 | 493 | 11 | 0 | 19 | |
| GSE43470 | ATP2B4 | 493 | 6 | 0 | 37 | |
| GSE46452 | ATP2B4 | 493 | 3 | 1 | 55 | |
| GSE47630 | ATP2B4 | 493 | 14 | 0 | 26 | |
| GSE54993 | ATP2B4 | 493 | 0 | 6 | 64 | |
| GSE54994 | ATP2B4 | 493 | 15 | 0 | 38 | |
| GSE60625 | ATP2B4 | 493 | 0 | 0 | 11 | |
| GSE74703 | ATP2B4 | 493 | 6 | 0 | 30 | |
| GSE74704 | ATP2B4 | 493 | 5 | 0 | 15 | |
| TCGA | ATP2B4 | 493 | 44 | 4 | 48 |
Total number of gains: 115; Total number of losses: 11; Total Number of normals: 362.
Somatic mutations of ATP2B4:
Generating mutation plots.
Highly correlated genes for ATP2B4:
Showing top 20/755 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP2B4 | FERMT2 | 0.817012 | 5 | 0 | 5 |
| ATP2B4 | TMTC1 | 0.797188 | 3 | 0 | 3 |
| ATP2B4 | ABL1 | 0.759141 | 5 | 0 | 5 |
| ATP2B4 | FOXN3 | 0.758893 | 3 | 0 | 3 |
| ATP2B4 | CCDC136 | 0.755177 | 3 | 0 | 3 |
| ATP2B4 | INMT | 0.755017 | 3 | 0 | 3 |
| ATP2B4 | TCEA2 | 0.744174 | 3 | 0 | 3 |
| ATP2B4 | FNDC5 | 0.737714 | 3 | 0 | 3 |
| ATP2B4 | SNRNP70 | 0.73226 | 3 | 0 | 3 |
| ATP2B4 | NCAM1 | 0.732033 | 5 | 0 | 4 |
| ATP2B4 | LAMA4 | 0.730804 | 5 | 0 | 4 |
| ATP2B4 | SGCB | 0.725993 | 5 | 0 | 5 |
| ATP2B4 | TAF6 | 0.713747 | 3 | 0 | 3 |
| ATP2B4 | MAP1B | 0.712961 | 7 | 0 | 5 |
| ATP2B4 | TSPAN18 | 0.70947 | 4 | 0 | 4 |
| ATP2B4 | PARVA | 0.706658 | 5 | 0 | 4 |
| ATP2B4 | EML1 | 0.704453 | 6 | 0 | 5 |
| ATP2B4 | S1PR3 | 0.704106 | 4 | 0 | 4 |
| ATP2B4 | PGR | 0.700631 | 3 | 0 | 3 |
| ATP2B4 | ILK | 0.699897 | 7 | 0 | 5 |
For details and further investigation, click here