| Full name: Pvt1 oncogene | Alias Symbol: NCRNA00079|LINC00079|onco-lncRNA-100|MIR1204HG | ||
| Type: non-coding RNA | Cytoband: 8q24.21 | ||
| Entrez ID: 5820 | HGNC ID: HGNC:9709 | Ensembl Gene: ENSG00000249859 | OMIM ID: 165140 |
| Drug and gene relationship at DGIdb | |||
Expression of PVT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | PVT1 | 5820 | 118575 | 0.8299 | 0.0000 | |
| GSE53624 | PVT1 | 5820 | 118575 | 0.5895 | 0.0000 | |
| GSE97050 | PVT1 | A_21_P0005951 | 0.9871 | 0.1400 | ||
| SRP007169 | PVT1 | 5820 | RNAseq | 0.0886 | 0.8438 | |
| SRP008496 | PVT1 | 5820 | RNAseq | 0.1712 | 0.5805 | |
| SRP064894 | PVT1 | 5820 | RNAseq | 1.1518 | 0.0000 | |
| SRP133303 | PVT1 | 5820 | RNAseq | 0.5672 | 0.0049 | |
| SRP159526 | PVT1 | 5820 | RNAseq | 0.5918 | 0.0082 | |
| SRP193095 | PVT1 | 5820 | RNAseq | 0.8353 | 0.0000 | |
| SRP219564 | PVT1 | 5820 | RNAseq | 0.9167 | 0.0665 | |
| TCGA | PVT1 | 5820 | RNAseq | 1.1632 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 0.
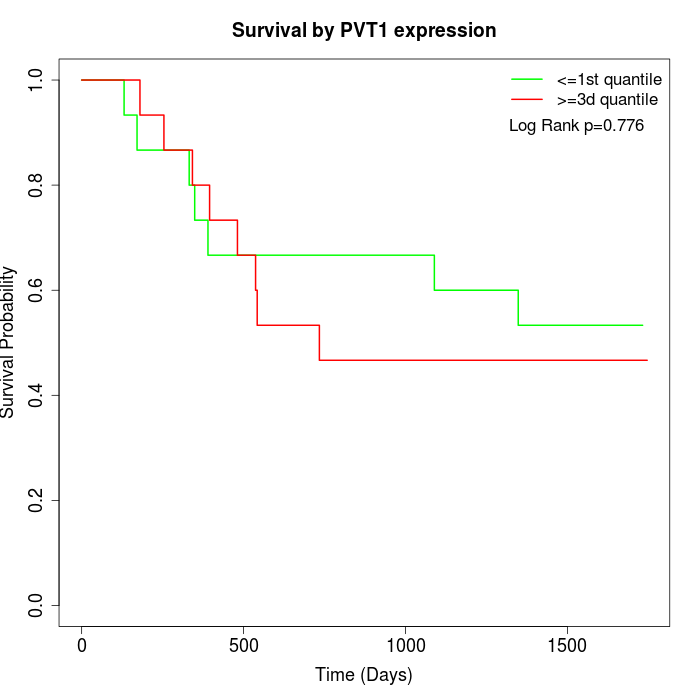
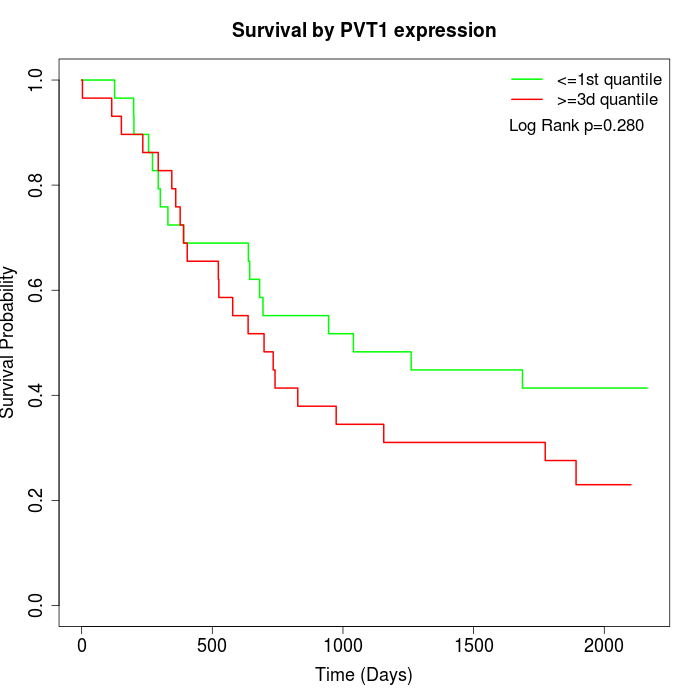
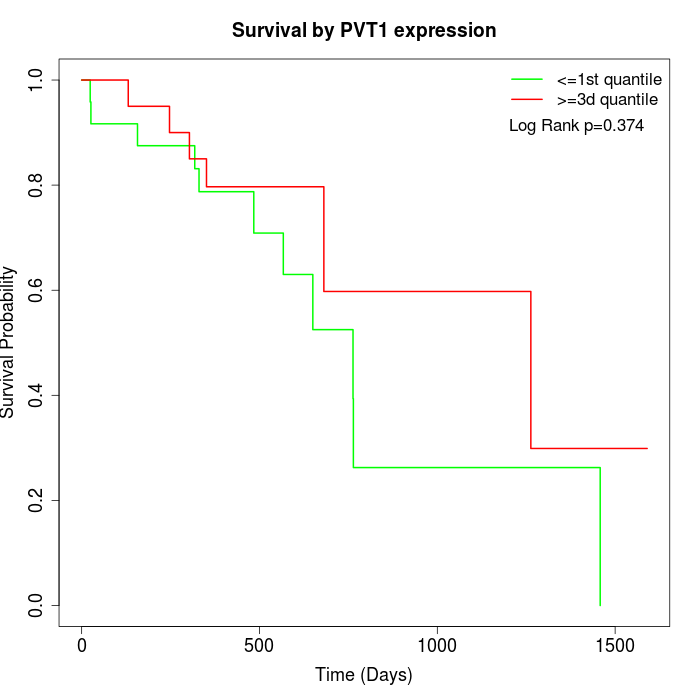
Survival by PVT1 expression:
Note: Click image to view full size file.
Copy number change of PVT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PVT1 | 5820 | 21 | 0 | 9 | |
| GSE20123 | PVT1 | 5820 | 21 | 0 | 9 | |
| GSE43470 | PVT1 | 5820 | 23 | 0 | 20 | |
| GSE46452 | PVT1 | 5820 | 30 | 0 | 29 | |
| GSE47630 | PVT1 | 5820 | 24 | 0 | 16 | |
| GSE54993 | PVT1 | 5820 | 1 | 24 | 45 | |
| GSE54994 | PVT1 | 5820 | 41 | 0 | 12 | |
| GSE60625 | PVT1 | 5820 | 0 | 7 | 4 | |
| GSE74703 | PVT1 | 5820 | 19 | 0 | 17 | |
| GSE74704 | PVT1 | 5820 | 14 | 0 | 6 | |
| TCGA | PVT1 | 5820 | 69 | 2 | 25 |
Total number of gains: 263; Total number of losses: 33; Total Number of normals: 192.
Somatic mutations of PVT1:
Generating mutation plots.
Highly correlated genes for PVT1:
Showing top 20/88 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PVT1 | MTIF2 | 0.731654 | 3 | 0 | 3 |
| PVT1 | TOP2A | 0.729764 | 3 | 0 | 3 |
| PVT1 | CDCA7 | 0.719162 | 3 | 0 | 3 |
| PVT1 | TROAP | 0.71133 | 3 | 0 | 3 |
| PVT1 | RAVER1 | 0.701872 | 3 | 0 | 3 |
| PVT1 | ATXN2L | 0.696788 | 3 | 0 | 3 |
| PVT1 | TAS2R43 | 0.695063 | 3 | 0 | 3 |
| PVT1 | MAZ | 0.691733 | 3 | 0 | 3 |
| PVT1 | SLC19A1 | 0.689326 | 3 | 0 | 3 |
| PVT1 | RBBP5 | 0.684687 | 3 | 0 | 3 |
| PVT1 | SLC38A6 | 0.677376 | 3 | 0 | 3 |
| PVT1 | ASPM | 0.674116 | 3 | 0 | 3 |
| PVT1 | MYBL2 | 0.674001 | 3 | 0 | 3 |
| PVT1 | SLC12A8 | 0.670035 | 3 | 0 | 3 |
| PVT1 | LMNB2 | 0.669181 | 3 | 0 | 3 |
| PVT1 | FAM111B | 0.667089 | 3 | 0 | 3 |
| PVT1 | SPDYE3 | 0.665942 | 3 | 0 | 3 |
| PVT1 | LEPR | 0.665542 | 3 | 0 | 3 |
| PVT1 | HELLS | 0.664524 | 3 | 0 | 3 |
| PVT1 | SPDYE1 | 0.663579 | 3 | 0 | 3 |
For details and further investigation, click here