| Full name: lamin B2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 84823 | HGNC ID: HGNC:6638 | Ensembl Gene: ENSG00000176619 | OMIM ID: 150341 |
| Drug and gene relationship at DGIdb | |||
LMNB2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04210 | Apoptosis |
Expression of LMNB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LMNB2 | 84823 | 216952_s_at | 1.4961 | 0.0174 | |
| GSE20347 | LMNB2 | 84823 | 216952_s_at | 0.8597 | 0.0002 | |
| GSE23400 | LMNB2 | 84823 | 216952_s_at | 0.9129 | 0.0000 | |
| GSE26886 | LMNB2 | 84823 | 216952_s_at | -0.2270 | 0.3277 | |
| GSE29001 | LMNB2 | 84823 | 216952_s_at | 1.0361 | 0.0054 | |
| GSE38129 | LMNB2 | 84823 | 216952_s_at | 1.3244 | 0.0000 | |
| GSE45670 | LMNB2 | 84823 | 216952_s_at | 1.1677 | 0.0000 | |
| GSE53622 | LMNB2 | 84823 | 91808 | 0.6711 | 0.0000 | |
| GSE53624 | LMNB2 | 84823 | 91808 | 0.5381 | 0.0000 | |
| GSE63941 | LMNB2 | 84823 | 216952_s_at | 0.9367 | 0.0039 | |
| GSE77861 | LMNB2 | 84823 | 216952_s_at | 1.1829 | 0.0011 | |
| GSE97050 | LMNB2 | 84823 | A_23_P67725 | 1.3319 | 0.0555 | |
| SRP007169 | LMNB2 | 84823 | RNAseq | 2.9453 | 0.0000 | |
| SRP008496 | LMNB2 | 84823 | RNAseq | 2.4524 | 0.0000 | |
| SRP064894 | LMNB2 | 84823 | RNAseq | 1.0899 | 0.0000 | |
| SRP133303 | LMNB2 | 84823 | RNAseq | 1.1977 | 0.0000 | |
| SRP159526 | LMNB2 | 84823 | RNAseq | 1.1837 | 0.0000 | |
| SRP193095 | LMNB2 | 84823 | RNAseq | 0.8988 | 0.0000 | |
| SRP219564 | LMNB2 | 84823 | RNAseq | 1.0870 | 0.0197 | |
| TCGA | LMNB2 | 84823 | RNAseq | 0.4943 | 0.0000 |
Upregulated datasets: 11; Downregulated datasets: 0.
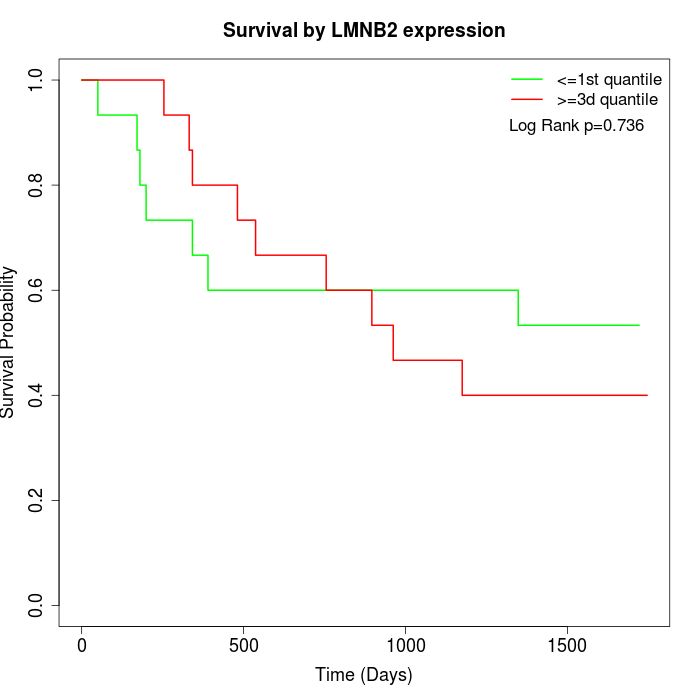
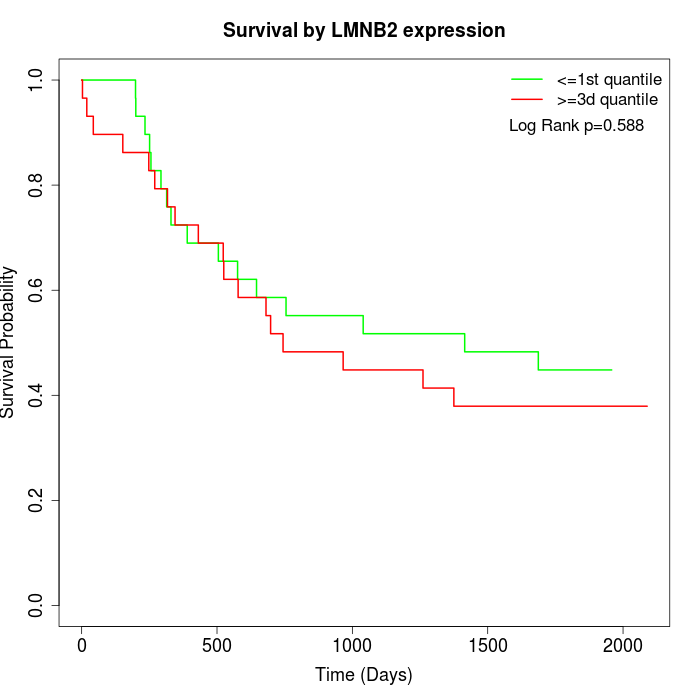
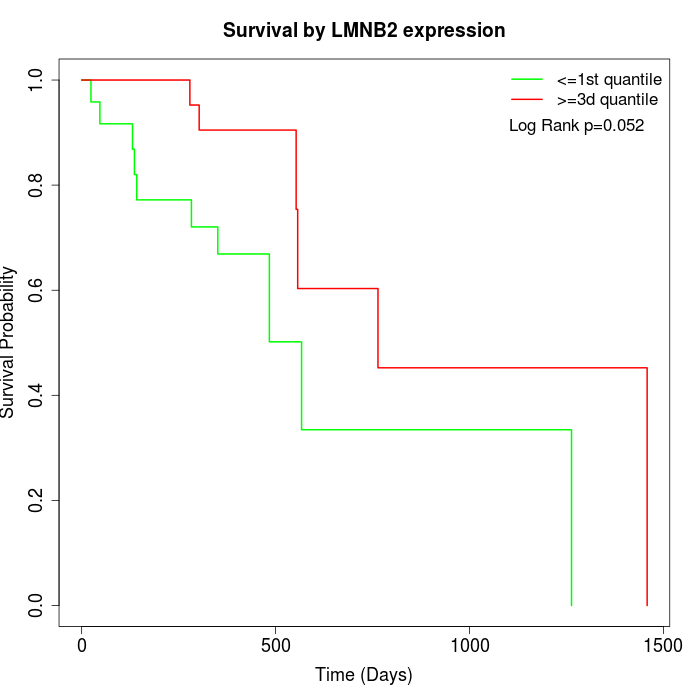
Survival by LMNB2 expression:
Note: Click image to view full size file.
Copy number change of LMNB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LMNB2 | 84823 | 4 | 4 | 22 | |
| GSE20123 | LMNB2 | 84823 | 4 | 4 | 22 | |
| GSE43470 | LMNB2 | 84823 | 1 | 10 | 32 | |
| GSE46452 | LMNB2 | 84823 | 47 | 1 | 11 | |
| GSE47630 | LMNB2 | 84823 | 5 | 7 | 28 | |
| GSE54993 | LMNB2 | 84823 | 16 | 3 | 51 | |
| GSE54994 | LMNB2 | 84823 | 8 | 15 | 30 | |
| GSE60625 | LMNB2 | 84823 | 9 | 0 | 2 | |
| GSE74703 | LMNB2 | 84823 | 1 | 7 | 28 | |
| GSE74704 | LMNB2 | 84823 | 2 | 3 | 15 | |
| TCGA | LMNB2 | 84823 | 6 | 25 | 65 |
Total number of gains: 103; Total number of losses: 79; Total Number of normals: 306.
Somatic mutations of LMNB2:
Generating mutation plots.
Highly correlated genes for LMNB2:
Showing top 20/1956 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LMNB2 | DDX39A | 0.7959 | 11 | 0 | 11 |
| LMNB2 | KHSRP | 0.788803 | 7 | 0 | 7 |
| LMNB2 | KPNA2 | 0.784949 | 8 | 0 | 8 |
| LMNB2 | LSM4 | 0.778267 | 10 | 0 | 10 |
| LMNB2 | CDCA3 | 0.777488 | 11 | 0 | 10 |
| LMNB2 | TTK | 0.777394 | 10 | 0 | 10 |
| LMNB2 | CENPF | 0.777231 | 12 | 0 | 10 |
| LMNB2 | MCM10 | 0.776385 | 10 | 0 | 9 |
| LMNB2 | DNMT1 | 0.772765 | 12 | 0 | 11 |
| LMNB2 | TMEM138 | 0.772549 | 5 | 0 | 5 |
| LMNB2 | XPR1 | 0.771124 | 6 | 0 | 6 |
| LMNB2 | CEP55 | 0.769535 | 13 | 0 | 12 |
| LMNB2 | RTKN | 0.769304 | 7 | 0 | 7 |
| LMNB2 | FZD6 | 0.769085 | 8 | 0 | 8 |
| LMNB2 | UBE2C | 0.768596 | 13 | 0 | 11 |
| LMNB2 | PSME4 | 0.766961 | 9 | 0 | 9 |
| LMNB2 | KNSTRN | 0.766583 | 6 | 0 | 6 |
| LMNB2 | PLXNA1 | 0.76385 | 10 | 0 | 10 |
| LMNB2 | NUF2 | 0.7616 | 8 | 0 | 7 |
| LMNB2 | SIX4 | 0.760346 | 6 | 0 | 6 |
For details and further investigation, click here