| Full name: PYD and CARD domain containing | Alias Symbol: TMS-1|CARD5|ASC | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 29108 | HGNC ID: HGNC:16608 | Ensembl Gene: ENSG00000103490 | OMIM ID: 606838 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PYCARD involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04621 | NOD-like receptor signaling pathway | |
| hsa05132 | Salmonella infection | |
| hsa05133 | Pertussis | |
| hsa05134 | Legionellosis | |
| hsa05164 | Influenza A |
Expression of PYCARD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PYCARD | 29108 | 221666_s_at | 0.3185 | 0.7286 | |
| GSE20347 | PYCARD | 29108 | 221666_s_at | -0.1725 | 0.5064 | |
| GSE23400 | PYCARD | 29108 | 221666_s_at | 0.6017 | 0.0001 | |
| GSE26886 | PYCARD | 29108 | 221666_s_at | 0.8221 | 0.0023 | |
| GSE29001 | PYCARD | 29108 | 221666_s_at | 0.2590 | 0.5555 | |
| GSE38129 | PYCARD | 29108 | 221666_s_at | 0.3092 | 0.1860 | |
| GSE45670 | PYCARD | 29108 | 221666_s_at | 0.6958 | 0.0009 | |
| GSE53622 | PYCARD | 29108 | 136021 | 0.6645 | 0.0000 | |
| GSE53624 | PYCARD | 29108 | 136021 | 0.7318 | 0.0000 | |
| GSE63941 | PYCARD | 29108 | 221666_s_at | -0.7203 | 0.7078 | |
| GSE77861 | PYCARD | 29108 | 221666_s_at | 0.1882 | 0.7273 | |
| GSE97050 | PYCARD | 29108 | A_23_P26629 | 0.6975 | 0.1867 | |
| SRP007169 | PYCARD | 29108 | RNAseq | -0.7800 | 0.1300 | |
| SRP008496 | PYCARD | 29108 | RNAseq | -0.4548 | 0.1529 | |
| SRP064894 | PYCARD | 29108 | RNAseq | 0.8658 | 0.0288 | |
| SRP133303 | PYCARD | 29108 | RNAseq | 0.4002 | 0.1233 | |
| SRP159526 | PYCARD | 29108 | RNAseq | 0.3018 | 0.4801 | |
| SRP193095 | PYCARD | 29108 | RNAseq | 0.0041 | 0.9821 | |
| SRP219564 | PYCARD | 29108 | RNAseq | 0.6211 | 0.1416 | |
| TCGA | PYCARD | 29108 | RNAseq | 0.6707 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
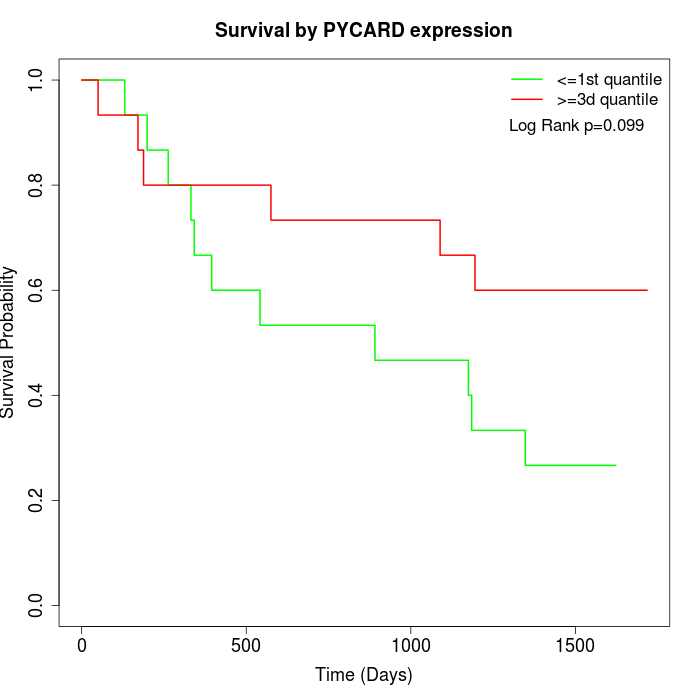
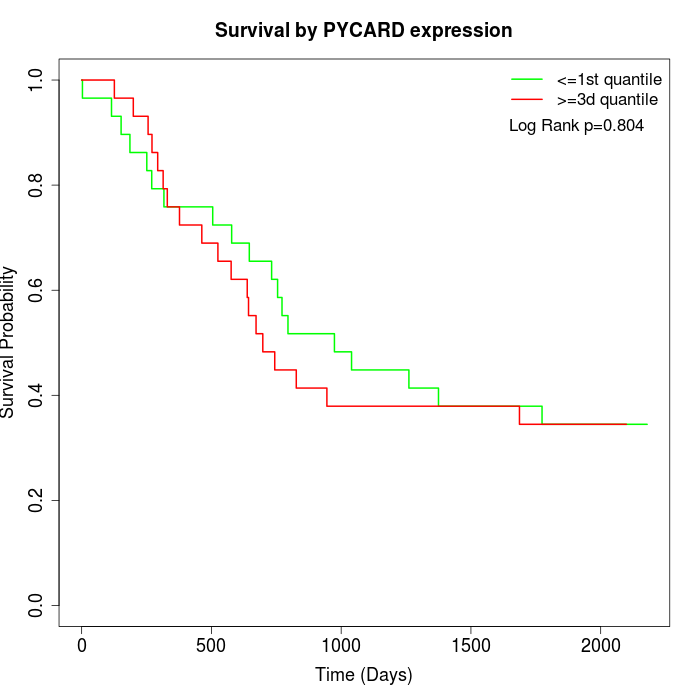
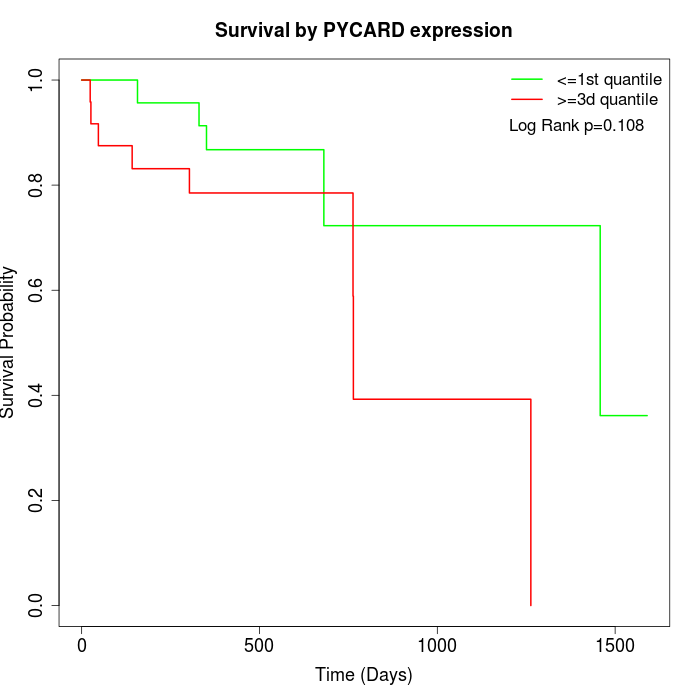
Survival by PYCARD expression:
Note: Click image to view full size file.
Copy number change of PYCARD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYCARD | 29108 | 4 | 5 | 21 | |
| GSE20123 | PYCARD | 29108 | 4 | 4 | 22 | |
| GSE43470 | PYCARD | 29108 | 3 | 3 | 37 | |
| GSE46452 | PYCARD | 29108 | 38 | 1 | 20 | |
| GSE47630 | PYCARD | 29108 | 11 | 7 | 22 | |
| GSE54993 | PYCARD | 29108 | 2 | 5 | 63 | |
| GSE54994 | PYCARD | 29108 | 5 | 9 | 39 | |
| GSE60625 | PYCARD | 29108 | 4 | 0 | 7 | |
| GSE74703 | PYCARD | 29108 | 3 | 2 | 31 | |
| GSE74704 | PYCARD | 29108 | 2 | 2 | 16 | |
| TCGA | PYCARD | 29108 | 20 | 10 | 66 |
Total number of gains: 96; Total number of losses: 48; Total Number of normals: 344.
Somatic mutations of PYCARD:
Generating mutation plots.
Highly correlated genes for PYCARD:
Showing top 20/584 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYCARD | PTTG2 | 0.701867 | 3 | 0 | 3 |
| PYCARD | ESCO2 | 0.69822 | 4 | 0 | 4 |
| PYCARD | EME1 | 0.69809 | 4 | 0 | 4 |
| PYCARD | MICAL1 | 0.68339 | 4 | 0 | 4 |
| PYCARD | SRXN1 | 0.670778 | 3 | 0 | 3 |
| PYCARD | GTSE1 | 0.666494 | 5 | 0 | 5 |
| PYCARD | TENM2 | 0.664473 | 5 | 0 | 4 |
| PYCARD | CCNB2 | 0.66404 | 8 | 0 | 7 |
| PYCARD | KRTDAP | 0.650868 | 4 | 0 | 3 |
| PYCARD | C12orf75 | 0.65014 | 6 | 0 | 5 |
| PYCARD | ADCK2 | 0.649639 | 3 | 0 | 3 |
| PYCARD | PPP1R14C | 0.646542 | 5 | 0 | 4 |
| PYCARD | FAM83B | 0.6435 | 4 | 0 | 3 |
| PYCARD | CD109 | 0.637066 | 3 | 0 | 3 |
| PYCARD | CCNA2 | 0.636974 | 7 | 0 | 5 |
| PYCARD | CCNF | 0.636031 | 7 | 0 | 6 |
| PYCARD | GEMIN4 | 0.633924 | 4 | 0 | 3 |
| PYCARD | G6PC3 | 0.632397 | 3 | 0 | 3 |
| PYCARD | CHSY1 | 0.631953 | 4 | 0 | 3 |
| PYCARD | MOV10 | 0.631572 | 3 | 0 | 3 |
For details and further investigation, click here