| Full name: cyclin F | Alias Symbol: FBX1|FBXO1 | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 899 | HGNC ID: HGNC:1591 | Ensembl Gene: ENSG00000162063 | OMIM ID: 600227 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CCNF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCNF | 899 | 204826_at | 0.8758 | 0.0378 | |
| GSE20347 | CCNF | 899 | 204826_at | 0.5828 | 0.0000 | |
| GSE23400 | CCNF | 899 | 204826_at | 0.6937 | 0.0000 | |
| GSE26886 | CCNF | 899 | 204826_at | 0.4008 | 0.0351 | |
| GSE29001 | CCNF | 899 | 204826_at | 0.8087 | 0.0016 | |
| GSE38129 | CCNF | 899 | 204826_at | 0.9461 | 0.0000 | |
| GSE45670 | CCNF | 899 | 204826_at | 0.9365 | 0.0000 | |
| GSE53622 | CCNF | 899 | 57367 | 1.7226 | 0.0000 | |
| GSE53624 | CCNF | 899 | 57367 | 1.6360 | 0.0000 | |
| GSE63941 | CCNF | 899 | 204826_at | 1.1534 | 0.0070 | |
| GSE77861 | CCNF | 899 | 204826_at | 0.1668 | 0.2616 | |
| GSE97050 | CCNF | 899 | A_24_P193592 | 1.0866 | 0.1985 | |
| SRP007169 | CCNF | 899 | RNAseq | 1.6887 | 0.0004 | |
| SRP008496 | CCNF | 899 | RNAseq | 1.0242 | 0.0037 | |
| SRP064894 | CCNF | 899 | RNAseq | 1.2509 | 0.0000 | |
| SRP133303 | CCNF | 899 | RNAseq | 1.4942 | 0.0000 | |
| SRP159526 | CCNF | 899 | RNAseq | 1.3501 | 0.0001 | |
| SRP193095 | CCNF | 899 | RNAseq | 1.0436 | 0.0000 | |
| SRP219564 | CCNF | 899 | RNAseq | 0.9413 | 0.1035 | |
| TCGA | CCNF | 899 | RNAseq | 0.8268 | 0.0000 |
Upregulated datasets: 9; Downregulated datasets: 0.
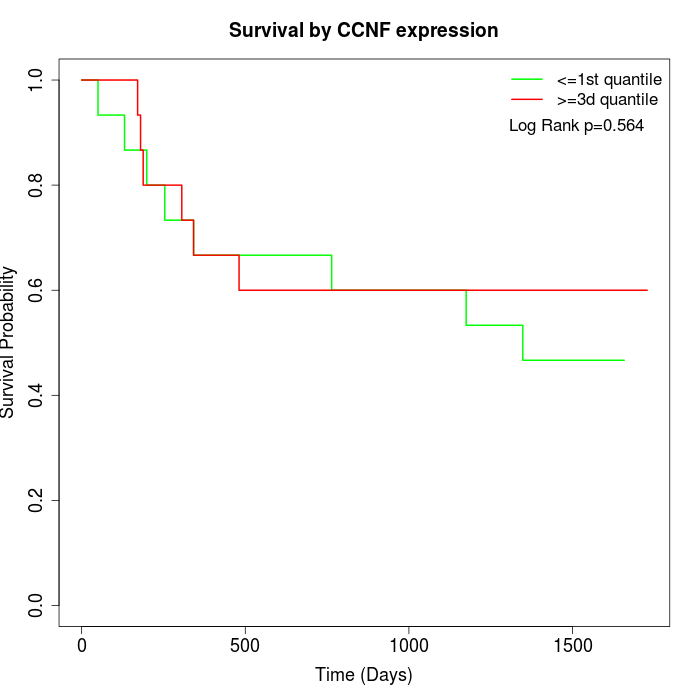
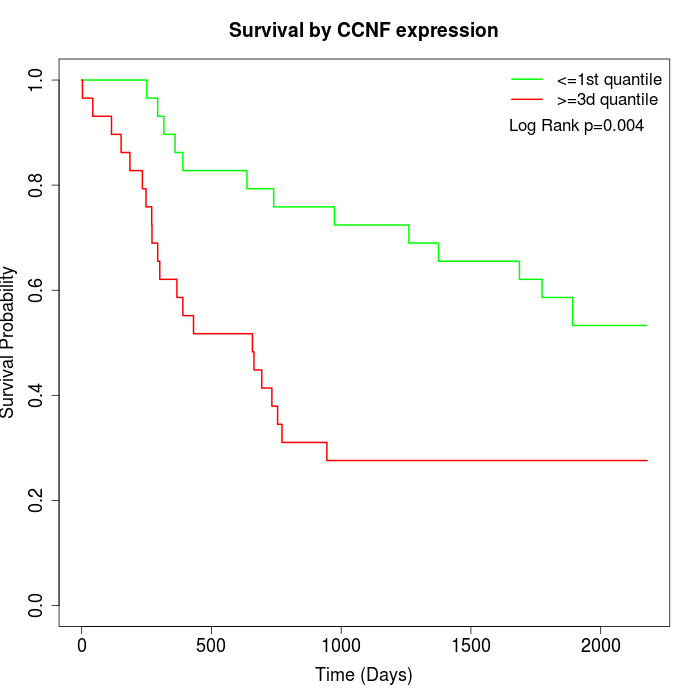
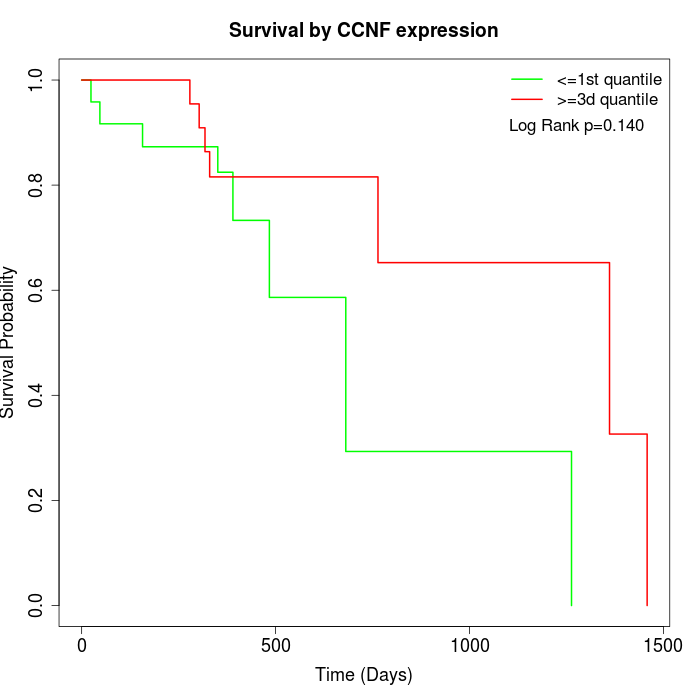
Survival by CCNF expression:
Note: Click image to view full size file.
Copy number change of CCNF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCNF | 899 | 5 | 5 | 20 | |
| GSE20123 | CCNF | 899 | 5 | 4 | 21 | |
| GSE43470 | CCNF | 899 | 4 | 7 | 32 | |
| GSE46452 | CCNF | 899 | 38 | 1 | 20 | |
| GSE47630 | CCNF | 899 | 13 | 6 | 21 | |
| GSE54993 | CCNF | 899 | 3 | 5 | 62 | |
| GSE54994 | CCNF | 899 | 5 | 9 | 39 | |
| GSE60625 | CCNF | 899 | 4 | 0 | 7 | |
| GSE74703 | CCNF | 899 | 4 | 5 | 27 | |
| GSE74704 | CCNF | 899 | 3 | 2 | 15 | |
| TCGA | CCNF | 899 | 20 | 13 | 63 |
Total number of gains: 104; Total number of losses: 57; Total Number of normals: 327.
Somatic mutations of CCNF:
Generating mutation plots.
Highly correlated genes for CCNF:
Showing top 20/1887 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCNF | UCK2 | 0.805302 | 3 | 0 | 3 |
| CCNF | DTL | 0.798797 | 12 | 0 | 12 |
| CCNF | STIL | 0.796131 | 11 | 0 | 11 |
| CCNF | MMS22L | 0.796018 | 3 | 0 | 3 |
| CCNF | UBE2C | 0.792522 | 12 | 0 | 12 |
| CCNF | TPX2 | 0.789845 | 12 | 0 | 12 |
| CCNF | KIF2C | 0.789639 | 12 | 0 | 11 |
| CCNF | DKC1 | 0.788333 | 3 | 0 | 3 |
| CCNF | RACGAP1 | 0.787525 | 9 | 0 | 8 |
| CCNF | AURKA | 0.786928 | 12 | 0 | 11 |
| CCNF | GINS1 | 0.781953 | 12 | 0 | 12 |
| CCNF | CCNA2 | 0.777587 | 11 | 0 | 10 |
| CCNF | CEP55 | 0.774566 | 11 | 0 | 10 |
| CCNF | CENPN | 0.772695 | 11 | 0 | 10 |
| CCNF | WDHD1 | 0.772626 | 11 | 0 | 11 |
| CCNF | LRRC58 | 0.771368 | 3 | 0 | 3 |
| CCNF | ESCO2 | 0.76905 | 7 | 0 | 7 |
| CCNF | MYBL2 | 0.767665 | 12 | 0 | 12 |
| CCNF | BIRC5 | 0.767517 | 13 | 0 | 12 |
| CCNF | CENPW | 0.767193 | 7 | 0 | 6 |
For details and further investigation, click here