| Full name: RAS like proto-oncogene B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q14.2 | ||
| Entrez ID: 5899 | HGNC ID: HGNC:9840 | Ensembl Gene: ENSG00000144118 | OMIM ID: 179551 |
| Drug and gene relationship at DGIdb | |||
RALB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway | |
| hsa04015 | Rap1 signaling pathway | |
| hsa04072 | Phospholipase D signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05212 | Pancreatic cancer |
Expression of RALB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RALB | 5899 | 202100_at | -0.6221 | 0.2571 | |
| GSE20347 | RALB | 5899 | 202100_at | -0.7672 | 0.0000 | |
| GSE23400 | RALB | 5899 | 202100_at | -0.3764 | 0.0000 | |
| GSE26886 | RALB | 5899 | 202100_at | -1.5098 | 0.0000 | |
| GSE29001 | RALB | 5899 | 202100_at | -0.6825 | 0.0304 | |
| GSE38129 | RALB | 5899 | 202100_at | -0.4041 | 0.0535 | |
| GSE45670 | RALB | 5899 | 202100_at | 0.0158 | 0.9490 | |
| GSE53622 | RALB | 5899 | 20276 | -0.2600 | 0.0058 | |
| GSE53624 | RALB | 5899 | 40951 | -0.1881 | 0.0066 | |
| GSE63941 | RALB | 5899 | 202100_at | 0.1055 | 0.8728 | |
| GSE77861 | RALB | 5899 | 202100_at | -0.2414 | 0.5426 | |
| GSE97050 | RALB | 5899 | A_33_P3356502 | -0.0737 | 0.8299 | |
| SRP007169 | RALB | 5899 | RNAseq | -0.8117 | 0.0516 | |
| SRP008496 | RALB | 5899 | RNAseq | -0.5596 | 0.1049 | |
| SRP064894 | RALB | 5899 | RNAseq | -0.4485 | 0.0112 | |
| SRP133303 | RALB | 5899 | RNAseq | -0.0962 | 0.6472 | |
| SRP159526 | RALB | 5899 | RNAseq | -0.0539 | 0.9162 | |
| SRP193095 | RALB | 5899 | RNAseq | -0.4988 | 0.0042 | |
| SRP219564 | RALB | 5899 | RNAseq | -0.8408 | 0.0240 | |
| TCGA | RALB | 5899 | RNAseq | 0.1195 | 0.0903 |
Upregulated datasets: 0; Downregulated datasets: 1.
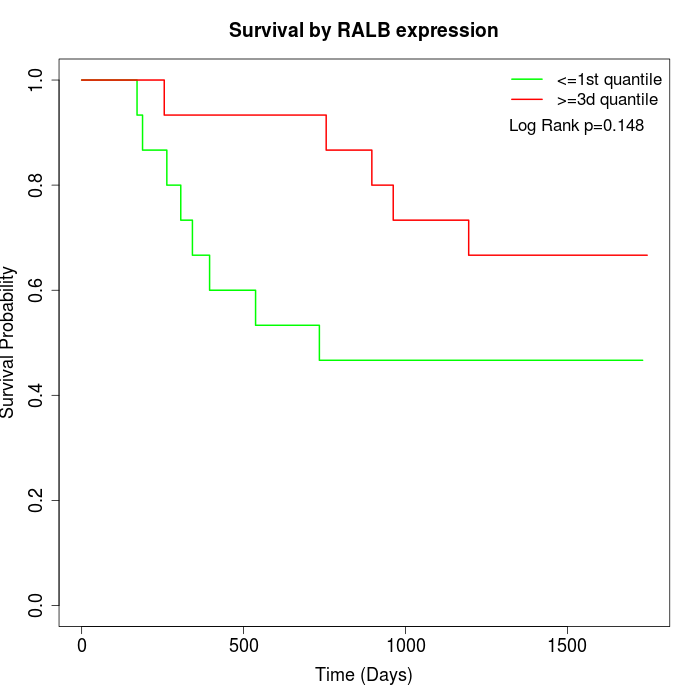
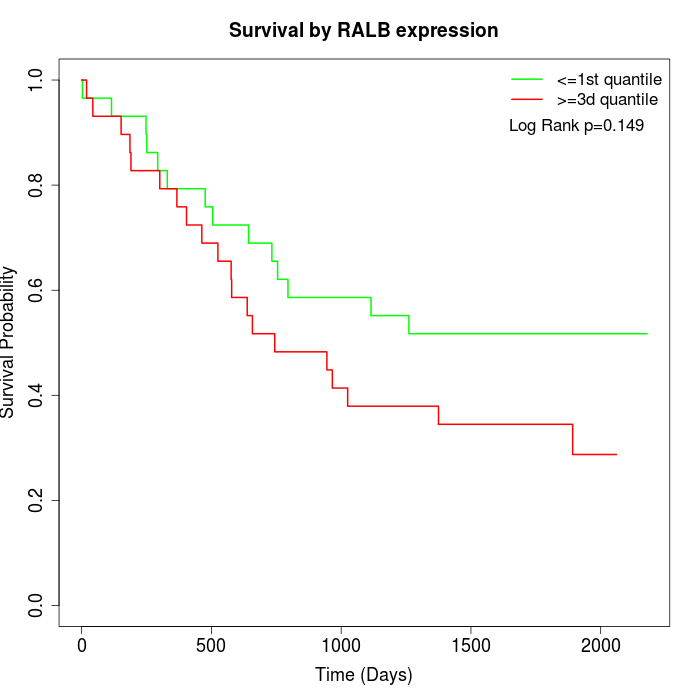
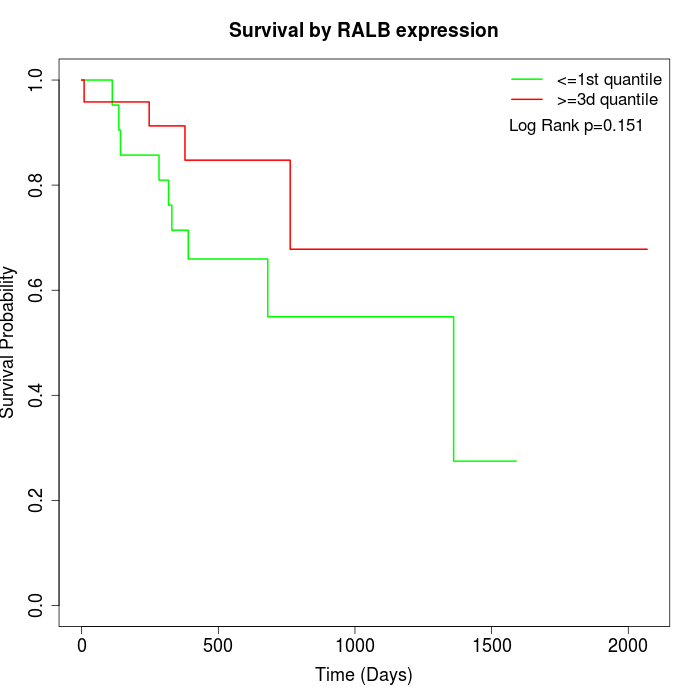
Survival by RALB expression:
Note: Click image to view full size file.
Copy number change of RALB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RALB | 5899 | 6 | 2 | 22 | |
| GSE20123 | RALB | 5899 | 6 | 2 | 22 | |
| GSE43470 | RALB | 5899 | 3 | 1 | 39 | |
| GSE46452 | RALB | 5899 | 2 | 3 | 54 | |
| GSE47630 | RALB | 5899 | 6 | 0 | 34 | |
| GSE54993 | RALB | 5899 | 0 | 6 | 64 | |
| GSE54994 | RALB | 5899 | 12 | 0 | 41 | |
| GSE60625 | RALB | 5899 | 0 | 3 | 8 | |
| GSE74703 | RALB | 5899 | 3 | 1 | 32 | |
| GSE74704 | RALB | 5899 | 3 | 1 | 16 | |
| TCGA | RALB | 5899 | 32 | 6 | 58 |
Total number of gains: 73; Total number of losses: 25; Total Number of normals: 390.
Somatic mutations of RALB:
Generating mutation plots.
Highly correlated genes for RALB:
Showing top 20/1178 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RALB | KCNK6 | 0.763142 | 4 | 0 | 4 |
| RALB | CAPNS2 | 0.761272 | 4 | 0 | 4 |
| RALB | LRRC28 | 0.737726 | 3 | 0 | 3 |
| RALB | GANC | 0.731145 | 4 | 0 | 4 |
| RALB | ULK3 | 0.728242 | 4 | 0 | 4 |
| RALB | C15orf62 | 0.727164 | 4 | 0 | 4 |
| RALB | MUC15 | 0.724019 | 3 | 0 | 3 |
| RALB | KIF21A | 0.715867 | 3 | 0 | 3 |
| RALB | SPTLC2 | 0.700607 | 9 | 0 | 8 |
| RALB | THAP2 | 0.698991 | 3 | 0 | 3 |
| RALB | XKRX | 0.697289 | 3 | 0 | 3 |
| RALB | ORMDL2 | 0.693684 | 8 | 0 | 7 |
| RALB | PHLDA1 | 0.693172 | 8 | 0 | 8 |
| RALB | FRMD4B | 0.691729 | 8 | 0 | 8 |
| RALB | PPP2R2A | 0.691207 | 8 | 0 | 8 |
| RALB | TOM1 | 0.69029 | 10 | 0 | 9 |
| RALB | MPZL3 | 0.68974 | 5 | 0 | 4 |
| RALB | OSTF1 | 0.689568 | 12 | 0 | 10 |
| RALB | UBE2D2 | 0.687491 | 5 | 0 | 4 |
| RALB | FLG-AS1 | 0.686105 | 3 | 0 | 3 |
For details and further investigation, click here