| Full name: ral guanine nucleotide dissociation stimulator | Alias Symbol: RGF|RalGEF|RGDS | ||
| Type: protein-coding gene | Cytoband: 9q34.13-q34.2 | ||
| Entrez ID: 5900 | HGNC ID: HGNC:9842 | Ensembl Gene: ENSG00000160271 | OMIM ID: 601619 |
| Drug and gene relationship at DGIdb | |||
RALGDS involved pathways:
Expression of RALGDS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RALGDS | 5900 | 209050_s_at | -0.1847 | 0.7273 | |
| GSE20347 | RALGDS | 5900 | 209050_s_at | -0.2951 | 0.1495 | |
| GSE23400 | RALGDS | 5900 | 209050_s_at | -0.0937 | 0.3073 | |
| GSE26886 | RALGDS | 5900 | 209050_s_at | 0.5202 | 0.0526 | |
| GSE29001 | RALGDS | 5900 | 209050_s_at | -0.2944 | 0.4599 | |
| GSE38129 | RALGDS | 5900 | 209050_s_at | -0.1321 | 0.4228 | |
| GSE45670 | RALGDS | 5900 | 209050_s_at | -0.2640 | 0.1291 | |
| GSE63941 | RALGDS | 5900 | 209050_s_at | 0.2020 | 0.7394 | |
| GSE77861 | RALGDS | 5900 | 209050_s_at | 0.5797 | 0.1221 | |
| GSE97050 | RALGDS | 5900 | A_23_P135184 | -0.1723 | 0.5008 | |
| SRP007169 | RALGDS | 5900 | RNAseq | 0.2433 | 0.4935 | |
| SRP008496 | RALGDS | 5900 | RNAseq | 0.1362 | 0.5392 | |
| SRP064894 | RALGDS | 5900 | RNAseq | -0.3461 | 0.1927 | |
| SRP133303 | RALGDS | 5900 | RNAseq | -0.3148 | 0.1118 | |
| SRP159526 | RALGDS | 5900 | RNAseq | -0.4339 | 0.1870 | |
| SRP193095 | RALGDS | 5900 | RNAseq | 0.0609 | 0.6677 | |
| SRP219564 | RALGDS | 5900 | RNAseq | -0.4967 | 0.1181 | |
| TCGA | RALGDS | 5900 | RNAseq | 0.0541 | 0.2931 |
Upregulated datasets: 0; Downregulated datasets: 0.
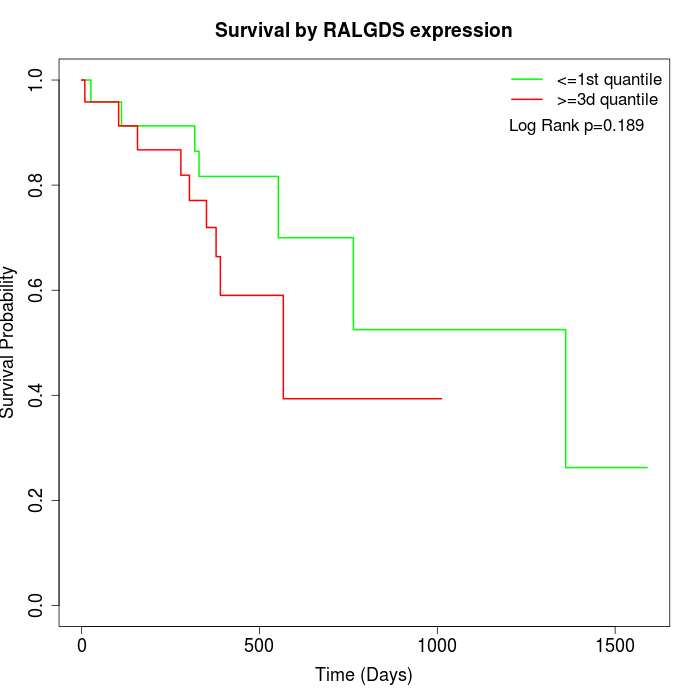
Survival by RALGDS expression:
Note: Click image to view full size file.
Copy number change of RALGDS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RALGDS | 5900 | 5 | 7 | 18 | |
| GSE20123 | RALGDS | 5900 | 5 | 7 | 18 | |
| GSE43470 | RALGDS | 5900 | 4 | 7 | 32 | |
| GSE46452 | RALGDS | 5900 | 6 | 13 | 40 | |
| GSE47630 | RALGDS | 5900 | 4 | 15 | 21 | |
| GSE54993 | RALGDS | 5900 | 3 | 3 | 64 | |
| GSE54994 | RALGDS | 5900 | 12 | 9 | 32 | |
| GSE60625 | RALGDS | 5900 | 0 | 0 | 11 | |
| GSE74703 | RALGDS | 5900 | 4 | 5 | 27 | |
| GSE74704 | RALGDS | 5900 | 3 | 5 | 12 | |
| TCGA | RALGDS | 5900 | 28 | 26 | 42 |
Total number of gains: 74; Total number of losses: 97; Total Number of normals: 317.
Somatic mutations of RALGDS:
Generating mutation plots.
Highly correlated genes for RALGDS:
Showing top 20/32 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RALGDS | LUZP1 | 0.722705 | 3 | 0 | 3 |
| RALGDS | PARP10 | 0.718639 | 3 | 0 | 3 |
| RALGDS | DOHH | 0.693407 | 4 | 0 | 4 |
| RALGDS | NOXA1 | 0.680179 | 3 | 0 | 3 |
| RALGDS | WDPCP | 0.66427 | 3 | 0 | 3 |
| RALGDS | FASTK | 0.652592 | 4 | 0 | 3 |
| RALGDS | FBXO44 | 0.650076 | 3 | 0 | 3 |
| RALGDS | ATG16L1 | 0.628404 | 3 | 0 | 3 |
| RALGDS | MC1R | 0.626761 | 3 | 0 | 3 |
| RALGDS | IRF3 | 0.604876 | 3 | 0 | 3 |
| RALGDS | EME2 | 0.603178 | 3 | 0 | 3 |
| RALGDS | KLHDC4 | 0.590725 | 3 | 0 | 3 |
| RALGDS | EVX1 | 0.584277 | 5 | 0 | 3 |
| RALGDS | ACAP3 | 0.575806 | 4 | 0 | 3 |
| RALGDS | HYAL1 | 0.568898 | 5 | 0 | 3 |
| RALGDS | ABHD11 | 0.561661 | 5 | 0 | 3 |
| RALGDS | ANKRD52 | 0.561426 | 3 | 0 | 3 |
| RALGDS | PPP1R13B | 0.55705 | 7 | 0 | 3 |
| RALGDS | LTB4R | 0.55003 | 5 | 0 | 3 |
| RALGDS | RGS12 | 0.548002 | 7 | 0 | 5 |
For details and further investigation, click here