| Full name: RALBP1 associated Eps domain containing 2 | Alias Symbol: POB1 | ||
| Type: protein-coding gene | Cytoband: Xp22.2 | ||
| Entrez ID: 9185 | HGNC ID: HGNC:9963 | Ensembl Gene: ENSG00000169891 | OMIM ID: 300317 |
| Drug and gene relationship at DGIdb | |||
Expression of REPS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | REPS2 | 9185 | 205645_at | 0.3234 | 0.5049 | |
| GSE20347 | REPS2 | 9185 | 205645_at | -0.0830 | 0.4190 | |
| GSE23400 | REPS2 | 9185 | 205645_at | -0.0335 | 0.3867 | |
| GSE26886 | REPS2 | 9185 | 205645_at | 0.9458 | 0.0000 | |
| GSE29001 | REPS2 | 9185 | 205645_at | 0.0226 | 0.9224 | |
| GSE38129 | REPS2 | 9185 | 205645_at | -0.1591 | 0.1501 | |
| GSE45670 | REPS2 | 9185 | 205645_at | -0.1087 | 0.4751 | |
| GSE53622 | REPS2 | 9185 | 18113 | -0.5566 | 0.0000 | |
| GSE53624 | REPS2 | 9185 | 18113 | -0.3649 | 0.0000 | |
| GSE63941 | REPS2 | 9185 | 205645_at | 1.4481 | 0.0194 | |
| GSE77861 | REPS2 | 9185 | 205645_at | -0.0449 | 0.6671 | |
| GSE97050 | REPS2 | 9185 | A_33_P3357470 | -0.5450 | 0.1934 | |
| SRP007169 | REPS2 | 9185 | RNAseq | -0.1896 | 0.7396 | |
| SRP008496 | REPS2 | 9185 | RNAseq | -0.1045 | 0.8025 | |
| SRP064894 | REPS2 | 9185 | RNAseq | 0.0243 | 0.9238 | |
| SRP133303 | REPS2 | 9185 | RNAseq | 0.1141 | 0.7452 | |
| SRP159526 | REPS2 | 9185 | RNAseq | 0.2149 | 0.4488 | |
| SRP193095 | REPS2 | 9185 | RNAseq | -0.5494 | 0.1363 | |
| SRP219564 | REPS2 | 9185 | RNAseq | -0.3438 | 0.5377 | |
| TCGA | REPS2 | 9185 | RNAseq | -1.0573 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 1.
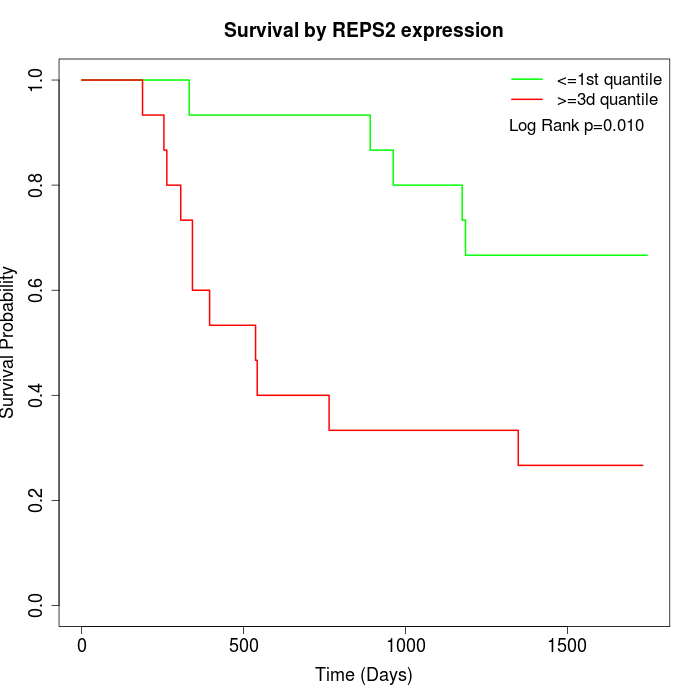
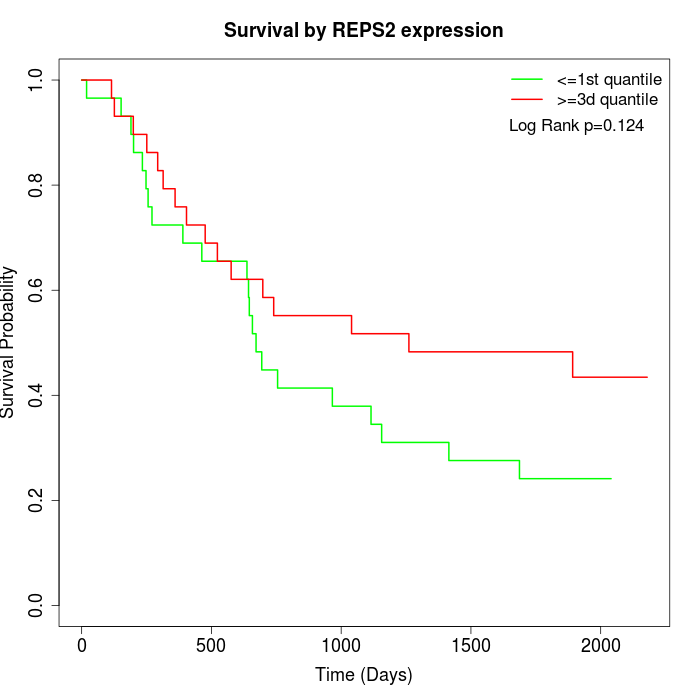
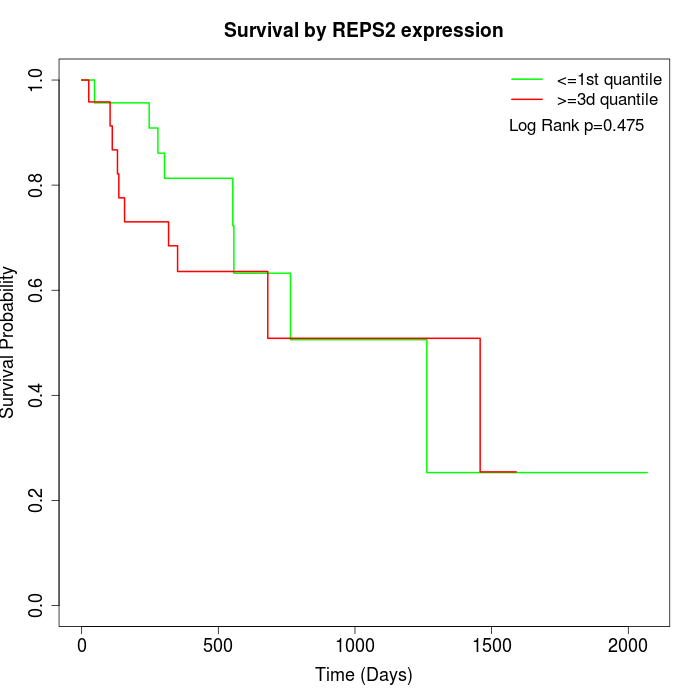
Survival by REPS2 expression:
Note: Click image to view full size file.
Copy number change of REPS2:
No record found for this gene.
Somatic mutations of REPS2:
Generating mutation plots.
Highly correlated genes for REPS2:
Showing top 20/132 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| REPS2 | SFXN5 | 0.757055 | 3 | 0 | 3 |
| REPS2 | TSNARE1 | 0.711816 | 3 | 0 | 3 |
| REPS2 | SYDE2 | 0.702054 | 3 | 0 | 3 |
| REPS2 | CHRM3 | 0.687942 | 3 | 0 | 3 |
| REPS2 | C2orf73 | 0.687051 | 4 | 0 | 4 |
| REPS2 | SIMC1 | 0.67477 | 3 | 0 | 3 |
| REPS2 | GKAP1 | 0.673603 | 4 | 0 | 3 |
| REPS2 | ADAMTSL3 | 0.668307 | 3 | 0 | 3 |
| REPS2 | FAM50B | 0.663037 | 3 | 0 | 3 |
| REPS2 | ISCU | 0.647488 | 5 | 0 | 4 |
| REPS2 | TAF5L | 0.647407 | 3 | 0 | 3 |
| REPS2 | LRRTM1 | 0.646921 | 4 | 0 | 3 |
| REPS2 | MRPL43 | 0.646529 | 4 | 0 | 4 |
| REPS2 | GATC | 0.646379 | 4 | 0 | 3 |
| REPS2 | NOTCH1 | 0.63708 | 4 | 0 | 4 |
| REPS2 | NAALADL2 | 0.636801 | 4 | 0 | 3 |
| REPS2 | WFDC3 | 0.634776 | 3 | 0 | 3 |
| REPS2 | ASB6 | 0.634049 | 3 | 0 | 3 |
| REPS2 | DZANK1 | 0.633707 | 5 | 0 | 4 |
| REPS2 | FOXO6 | 0.63002 | 4 | 0 | 3 |
For details and further investigation, click here