| Full name: ring finger protein 148 | Alias Symbol: MGC35222 | ||
| Type: protein-coding gene | Cytoband: 7q31.32 | ||
| Entrez ID: 378925 | HGNC ID: HGNC:22411 | Ensembl Gene: ENSG00000235631 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of RNF148:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RNF148 | 378925 | 242925_at | -0.1723 | 0.6051 | |
| GSE26886 | RNF148 | 378925 | 242925_at | -0.0712 | 0.4483 | |
| GSE45670 | RNF148 | 378925 | 242925_at | 0.1190 | 0.2450 | |
| GSE53622 | RNF148 | 378925 | 27244 | 0.2295 | 0.1920 | |
| GSE53624 | RNF148 | 378925 | 27244 | 0.3658 | 0.0043 | |
| GSE63941 | RNF148 | 378925 | 242925_at | -0.0484 | 0.7646 | |
| GSE77861 | RNF148 | 378925 | 242925_at | -0.1465 | 0.1005 | |
| GSE97050 | RNF148 | 378925 | A_24_P170234 | -0.1537 | 0.6443 | |
| TCGA | RNF148 | 378925 | RNAseq | -1.1092 | 0.0536 |
Upregulated datasets: 0; Downregulated datasets: 0.
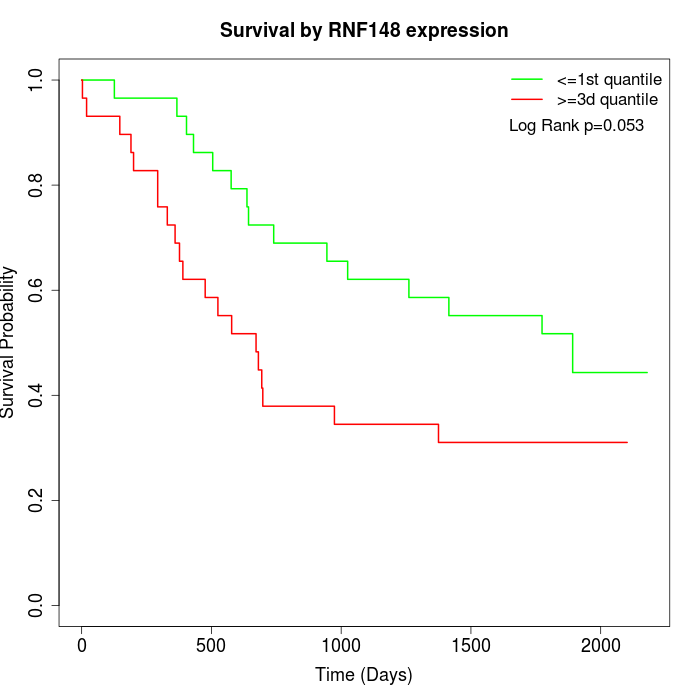
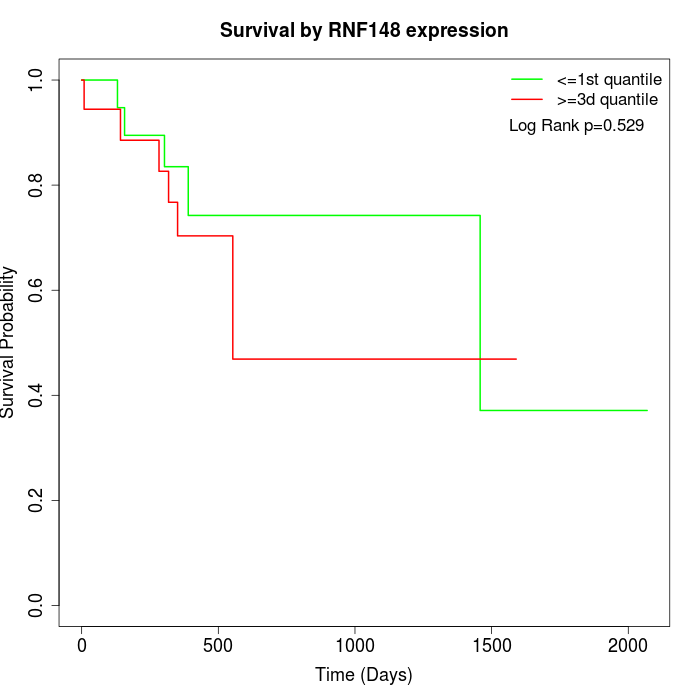
Survival by RNF148 expression:
Note: Click image to view full size file.
Copy number change of RNF148:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RNF148 | 378925 | 7 | 2 | 21 | |
| GSE20123 | RNF148 | 378925 | 8 | 2 | 20 | |
| GSE43470 | RNF148 | 378925 | 2 | 4 | 37 | |
| GSE46452 | RNF148 | 378925 | 8 | 1 | 50 | |
| GSE47630 | RNF148 | 378925 | 7 | 4 | 29 | |
| GSE54993 | RNF148 | 378925 | 3 | 5 | 62 | |
| GSE54994 | RNF148 | 378925 | 10 | 6 | 37 | |
| GSE60625 | RNF148 | 378925 | 0 | 0 | 11 | |
| GSE74703 | RNF148 | 378925 | 2 | 4 | 30 | |
| GSE74704 | RNF148 | 378925 | 5 | 2 | 13 | |
| TCGA | RNF148 | 378925 | 27 | 21 | 48 |
Total number of gains: 79; Total number of losses: 51; Total Number of normals: 358.
Somatic mutations of RNF148:
Generating mutation plots.
Highly correlated genes for RNF148:
Showing top 20/51 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RNF148 | CLYBL | 0.738785 | 3 | 0 | 3 |
| RNF148 | KCND3 | 0.723442 | 3 | 0 | 3 |
| RNF148 | ADAL | 0.720764 | 3 | 0 | 3 |
| RNF148 | ARRB1 | 0.719024 | 3 | 0 | 3 |
| RNF148 | PHYHIPL | 0.718957 | 3 | 0 | 3 |
| RNF148 | ITPRIP | 0.71139 | 3 | 0 | 3 |
| RNF148 | NKX6-1 | 0.708116 | 3 | 0 | 3 |
| RNF148 | SLC25A23 | 0.706881 | 3 | 0 | 3 |
| RNF148 | SRR | 0.704905 | 3 | 0 | 3 |
| RNF148 | SOX10 | 0.702246 | 3 | 0 | 3 |
| RNF148 | RNASEH2C | 0.683715 | 3 | 0 | 3 |
| RNF148 | SYN3 | 0.683558 | 3 | 0 | 3 |
| RNF148 | MYL3 | 0.665759 | 3 | 0 | 3 |
| RNF148 | CSRNP1 | 0.659779 | 3 | 0 | 3 |
| RNF148 | MAPK12 | 0.658026 | 3 | 0 | 3 |
| RNF148 | RND1 | 0.65471 | 3 | 0 | 3 |
| RNF148 | BTD | 0.651937 | 3 | 0 | 3 |
| RNF148 | HAS1 | 0.646629 | 3 | 0 | 3 |
| RNF148 | EIF4E3 | 0.639126 | 3 | 0 | 3 |
| RNF148 | FAM200B | 0.638554 | 3 | 0 | 3 |
For details and further investigation, click here