| Full name: SH3 domain binding protein 2 | Alias Symbol: RES4-23|CRBM | ||
| Type: protein-coding gene | Cytoband: 4p16.3 | ||
| Entrez ID: 6452 | HGNC ID: HGNC:10825 | Ensembl Gene: ENSG00000087266 | OMIM ID: 602104 |
| Drug and gene relationship at DGIdb | |||
SH3BP2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04650 | Natural killer cell mediated cytotoxicity |
Expression of SH3BP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SH3BP2 | 6452 | 209370_s_at | 0.3154 | 0.2187 | |
| GSE20347 | SH3BP2 | 6452 | 209370_s_at | -0.0507 | 0.6403 | |
| GSE23400 | SH3BP2 | 6452 | 209370_s_at | -0.1449 | 0.0001 | |
| GSE26886 | SH3BP2 | 6452 | 209370_s_at | -0.3088 | 0.0470 | |
| GSE29001 | SH3BP2 | 6452 | 209370_s_at | -0.2110 | 0.2283 | |
| GSE38129 | SH3BP2 | 6452 | 209370_s_at | -0.1570 | 0.1028 | |
| GSE45670 | SH3BP2 | 6452 | 209370_s_at | 0.2626 | 0.0063 | |
| GSE53622 | SH3BP2 | 6452 | 138150 | 0.2007 | 0.0283 | |
| GSE53624 | SH3BP2 | 6452 | 138150 | 0.2138 | 0.0050 | |
| GSE63941 | SH3BP2 | 6452 | 209370_s_at | -0.0989 | 0.6290 | |
| GSE77861 | SH3BP2 | 6452 | 209370_s_at | -0.0887 | 0.6054 | |
| GSE97050 | SH3BP2 | 6452 | A_33_P3414799 | 0.8140 | 0.1852 | |
| SRP007169 | SH3BP2 | 6452 | RNAseq | -0.6637 | 0.1040 | |
| SRP008496 | SH3BP2 | 6452 | RNAseq | -0.3880 | 0.1762 | |
| SRP064894 | SH3BP2 | 6452 | RNAseq | 0.2257 | 0.2689 | |
| SRP133303 | SH3BP2 | 6452 | RNAseq | -0.0377 | 0.8044 | |
| SRP159526 | SH3BP2 | 6452 | RNAseq | -0.1468 | 0.5944 | |
| SRP193095 | SH3BP2 | 6452 | RNAseq | 0.2098 | 0.1521 | |
| SRP219564 | SH3BP2 | 6452 | RNAseq | -0.1690 | 0.6855 | |
| TCGA | SH3BP2 | 6452 | RNAseq | -0.0584 | 0.3507 |
Upregulated datasets: 0; Downregulated datasets: 0.
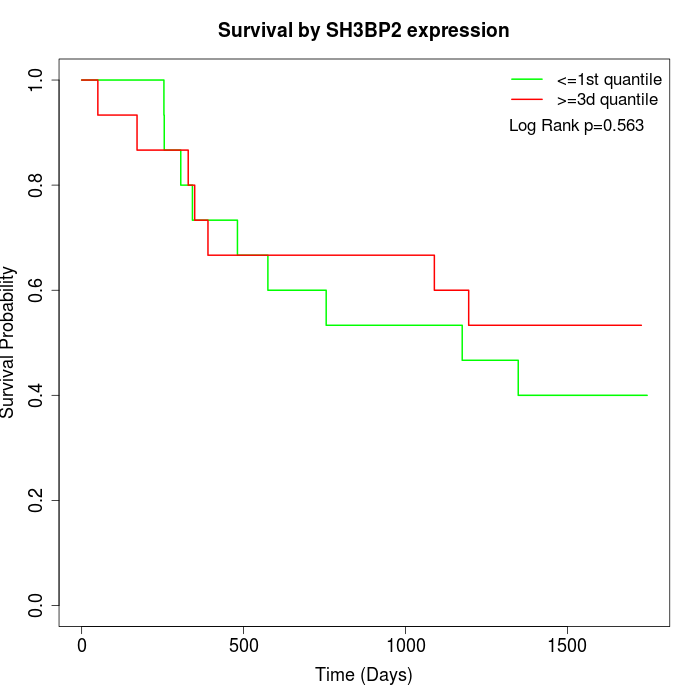
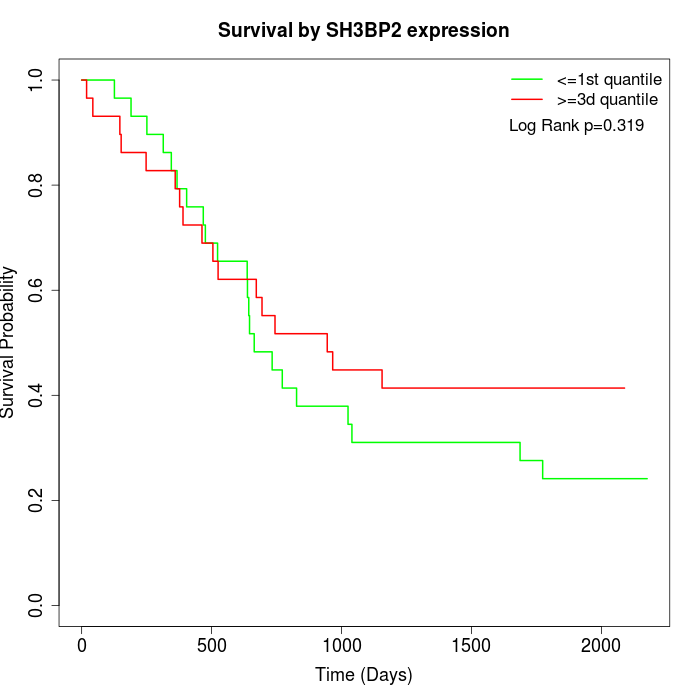
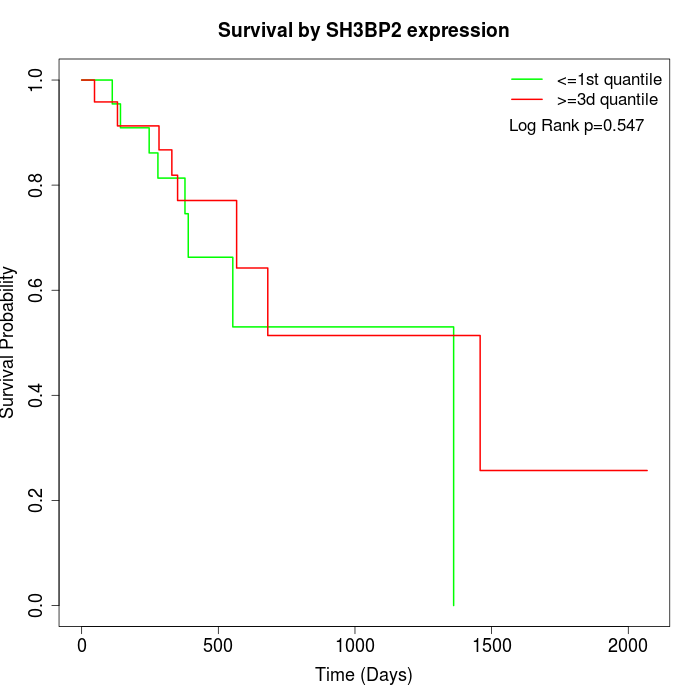
Survival by SH3BP2 expression:
Note: Click image to view full size file.
Copy number change of SH3BP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SH3BP2 | 6452 | 0 | 16 | 14 | |
| GSE20123 | SH3BP2 | 6452 | 0 | 16 | 14 | |
| GSE43470 | SH3BP2 | 6452 | 0 | 17 | 26 | |
| GSE46452 | SH3BP2 | 6452 | 1 | 36 | 22 | |
| GSE47630 | SH3BP2 | 6452 | 1 | 19 | 20 | |
| GSE54993 | SH3BP2 | 6452 | 11 | 0 | 59 | |
| GSE54994 | SH3BP2 | 6452 | 3 | 12 | 38 | |
| GSE60625 | SH3BP2 | 6452 | 0 | 0 | 11 | |
| GSE74703 | SH3BP2 | 6452 | 0 | 13 | 23 | |
| GSE74704 | SH3BP2 | 6452 | 0 | 8 | 12 | |
| TCGA | SH3BP2 | 6452 | 7 | 48 | 41 |
Total number of gains: 23; Total number of losses: 185; Total Number of normals: 280.
Somatic mutations of SH3BP2:
Generating mutation plots.
Highly correlated genes for SH3BP2:
Showing top 20/372 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SH3BP2 | LRRC56 | 0.763528 | 3 | 0 | 3 |
| SH3BP2 | MOCS3 | 0.760289 | 3 | 0 | 3 |
| SH3BP2 | C9orf16 | 0.743425 | 3 | 0 | 3 |
| SH3BP2 | FBRS | 0.739131 | 3 | 0 | 3 |
| SH3BP2 | SEMA4A | 0.732696 | 3 | 0 | 3 |
| SH3BP2 | APBA1 | 0.727943 | 4 | 0 | 4 |
| SH3BP2 | ZNF408 | 0.725076 | 5 | 0 | 5 |
| SH3BP2 | RAB11B | 0.722892 | 3 | 0 | 3 |
| SH3BP2 | FGF11 | 0.720757 | 3 | 0 | 3 |
| SH3BP2 | VAMP2 | 0.716125 | 3 | 0 | 3 |
| SH3BP2 | CORO1B | 0.712471 | 3 | 0 | 3 |
| SH3BP2 | EPN1 | 0.708841 | 5 | 0 | 4 |
| SH3BP2 | MOSPD3 | 0.708689 | 3 | 0 | 3 |
| SH3BP2 | RABEP2 | 0.706343 | 4 | 0 | 3 |
| SH3BP2 | CYP7B1 | 0.703621 | 3 | 0 | 3 |
| SH3BP2 | CCDC85C | 0.700219 | 4 | 0 | 3 |
| SH3BP2 | TNPO1 | 0.694133 | 3 | 0 | 3 |
| SH3BP2 | CLCN7 | 0.684591 | 4 | 0 | 4 |
| SH3BP2 | NMB | 0.683321 | 3 | 0 | 3 |
| SH3BP2 | MNT | 0.680574 | 3 | 0 | 3 |
For details and further investigation, click here