| Full name: SLIT-ROBO Rho GTPase activating protein 3 | Alias Symbol: KIAA0411|MEGAP|WRP|ARHGAP14 | ||
| Type: protein-coding gene | Cytoband: 3p25.3 | ||
| Entrez ID: 9901 | HGNC ID: HGNC:19744 | Ensembl Gene: ENSG00000196220 | OMIM ID: 606525 |
| Drug and gene relationship at DGIdb | |||
SRGAP3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of SRGAP3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SRGAP3 | 9901 | 209794_at | 0.0714 | 0.9284 | |
| GSE20347 | SRGAP3 | 9901 | 209794_at | -0.1416 | 0.2162 | |
| GSE23400 | SRGAP3 | 9901 | 209794_at | -0.0629 | 0.0752 | |
| GSE26886 | SRGAP3 | 9901 | 209794_at | -0.1517 | 0.4917 | |
| GSE29001 | SRGAP3 | 9901 | 209794_at | -0.3149 | 0.2387 | |
| GSE38129 | SRGAP3 | 9901 | 209794_at | -0.1016 | 0.3086 | |
| GSE45670 | SRGAP3 | 9901 | 209794_at | -0.0897 | 0.5970 | |
| GSE53622 | SRGAP3 | 9901 | 32389 | -0.2650 | 0.0209 | |
| GSE53624 | SRGAP3 | 9901 | 32389 | -0.4261 | 0.0000 | |
| GSE63941 | SRGAP3 | 9901 | 209794_at | 0.7161 | 0.3339 | |
| GSE77861 | SRGAP3 | 9901 | 209794_at | -0.0681 | 0.6370 | |
| GSE97050 | SRGAP3 | 9901 | A_23_P358410 | -0.0807 | 0.7715 | |
| SRP007169 | SRGAP3 | 9901 | RNAseq | 1.5471 | 0.0013 | |
| SRP064894 | SRGAP3 | 9901 | RNAseq | 0.0047 | 0.9892 | |
| SRP133303 | SRGAP3 | 9901 | RNAseq | -0.4451 | 0.1458 | |
| SRP159526 | SRGAP3 | 9901 | RNAseq | -0.1812 | 0.6560 | |
| SRP193095 | SRGAP3 | 9901 | RNAseq | -0.1393 | 0.5539 | |
| SRP219564 | SRGAP3 | 9901 | RNAseq | 0.0391 | 0.9213 | |
| TCGA | SRGAP3 | 9901 | RNAseq | -0.0373 | 0.7575 |
Upregulated datasets: 1; Downregulated datasets: 0.
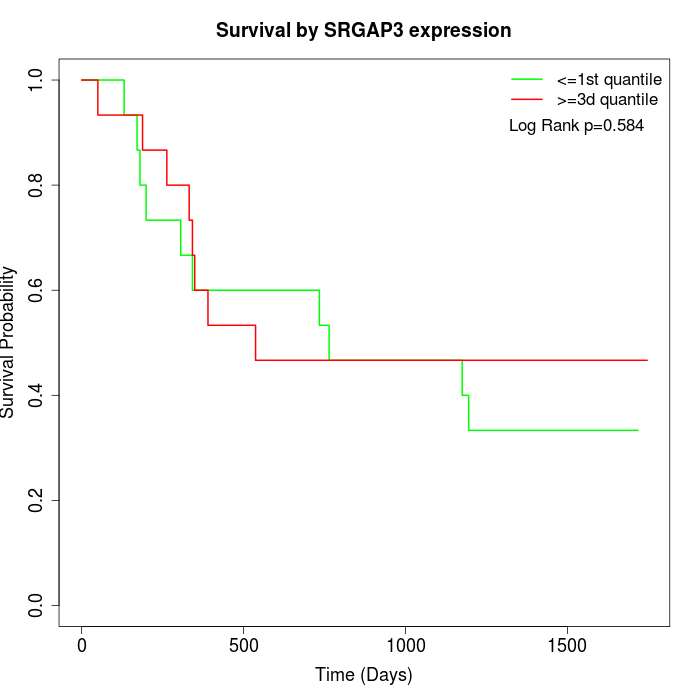
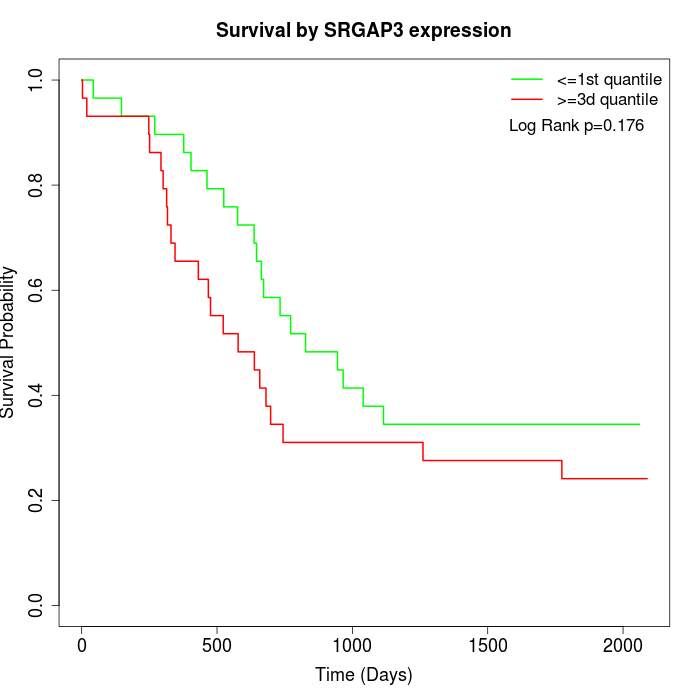
Survival by SRGAP3 expression:
Note: Click image to view full size file.
Copy number change of SRGAP3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SRGAP3 | 9901 | 1 | 18 | 11 | |
| GSE20123 | SRGAP3 | 9901 | 1 | 18 | 11 | |
| GSE43470 | SRGAP3 | 9901 | 0 | 21 | 22 | |
| GSE46452 | SRGAP3 | 9901 | 2 | 17 | 40 | |
| GSE47630 | SRGAP3 | 9901 | 2 | 22 | 16 | |
| GSE54993 | SRGAP3 | 9901 | 6 | 1 | 63 | |
| GSE54994 | SRGAP3 | 9901 | 3 | 31 | 19 | |
| GSE60625 | SRGAP3 | 9901 | 5 | 0 | 6 | |
| GSE74703 | SRGAP3 | 9901 | 0 | 17 | 19 | |
| GSE74704 | SRGAP3 | 9901 | 1 | 11 | 8 | |
| TCGA | SRGAP3 | 9901 | 6 | 64 | 26 |
Total number of gains: 27; Total number of losses: 220; Total Number of normals: 241.
Somatic mutations of SRGAP3:
Generating mutation plots.
Highly correlated genes for SRGAP3:
Showing top 20/153 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SRGAP3 | SMAD3 | 0.718013 | 3 | 0 | 3 |
| SRGAP3 | WDR77 | 0.710711 | 3 | 0 | 3 |
| SRGAP3 | CSK | 0.702789 | 3 | 0 | 3 |
| SRGAP3 | NFKBIL1 | 0.677516 | 3 | 0 | 3 |
| SRGAP3 | CCND1 | 0.677084 | 3 | 0 | 3 |
| SRGAP3 | KLHDC10 | 0.676628 | 3 | 0 | 3 |
| SRGAP3 | FBRS | 0.671728 | 3 | 0 | 3 |
| SRGAP3 | PRMT6 | 0.671409 | 3 | 0 | 3 |
| SRGAP3 | RSBN1L | 0.669958 | 3 | 0 | 3 |
| SRGAP3 | ALG6 | 0.667839 | 3 | 0 | 3 |
| SRGAP3 | COMMD2 | 0.657275 | 3 | 0 | 3 |
| SRGAP3 | E2F8 | 0.656408 | 4 | 0 | 3 |
| SRGAP3 | CTDSP1 | 0.655644 | 4 | 0 | 3 |
| SRGAP3 | ILDR1 | 0.651799 | 3 | 0 | 3 |
| SRGAP3 | CXCL16 | 0.641609 | 3 | 0 | 3 |
| SRGAP3 | GRAMD1C | 0.640772 | 4 | 0 | 3 |
| SRGAP3 | MRPL27 | 0.640296 | 4 | 0 | 4 |
| SRGAP3 | KHNYN | 0.637266 | 3 | 0 | 3 |
| SRGAP3 | ATG4C | 0.636318 | 3 | 0 | 3 |
| SRGAP3 | CD2 | 0.636028 | 3 | 0 | 3 |
For details and further investigation, click here