| Full name: sushi repeat containing protein X-linked 2 | Alias Symbol: SRPUL | ||
| Type: protein-coding gene | Cytoband: Xq22.1 | ||
| Entrez ID: 27286 | HGNC ID: HGNC:30668 | Ensembl Gene: ENSG00000102359 | OMIM ID: 300642 |
| Drug and gene relationship at DGIdb | |||
Expression of SRPX2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SRPX2 | 27286 | 205499_at | -0.7749 | 0.3743 | |
| GSE20347 | SRPX2 | 27286 | 205499_at | -1.0730 | 0.0010 | |
| GSE23400 | SRPX2 | 27286 | 205499_at | -0.5579 | 0.0000 | |
| GSE26886 | SRPX2 | 27286 | 205499_at | -2.6891 | 0.0000 | |
| GSE29001 | SRPX2 | 27286 | 205499_at | -1.8822 | 0.0025 | |
| GSE38129 | SRPX2 | 27286 | 205499_at | -0.5038 | 0.1033 | |
| GSE45670 | SRPX2 | 27286 | 205499_at | -0.4680 | 0.1086 | |
| GSE53622 | SRPX2 | 27286 | 62141 | -0.1872 | 0.1855 | |
| GSE53624 | SRPX2 | 27286 | 62141 | -0.8798 | 0.0000 | |
| GSE63941 | SRPX2 | 27286 | 205499_at | -2.6899 | 0.0491 | |
| GSE77861 | SRPX2 | 27286 | 205499_at | -0.7252 | 0.0540 | |
| GSE97050 | SRPX2 | 27286 | A_23_P136978 | 0.1750 | 0.7559 | |
| SRP007169 | SRPX2 | 27286 | RNAseq | -1.5906 | 0.0003 | |
| SRP008496 | SRPX2 | 27286 | RNAseq | -1.5227 | 0.0000 | |
| SRP064894 | SRPX2 | 27286 | RNAseq | -0.4488 | 0.0475 | |
| SRP133303 | SRPX2 | 27286 | RNAseq | 0.1663 | 0.5380 | |
| SRP159526 | SRPX2 | 27286 | RNAseq | -1.2509 | 0.0003 | |
| SRP193095 | SRPX2 | 27286 | RNAseq | -0.8244 | 0.0012 | |
| SRP219564 | SRPX2 | 27286 | RNAseq | -0.2392 | 0.7122 | |
| TCGA | SRPX2 | 27286 | RNAseq | 0.3656 | 0.0058 |
Upregulated datasets: 0; Downregulated datasets: 7.
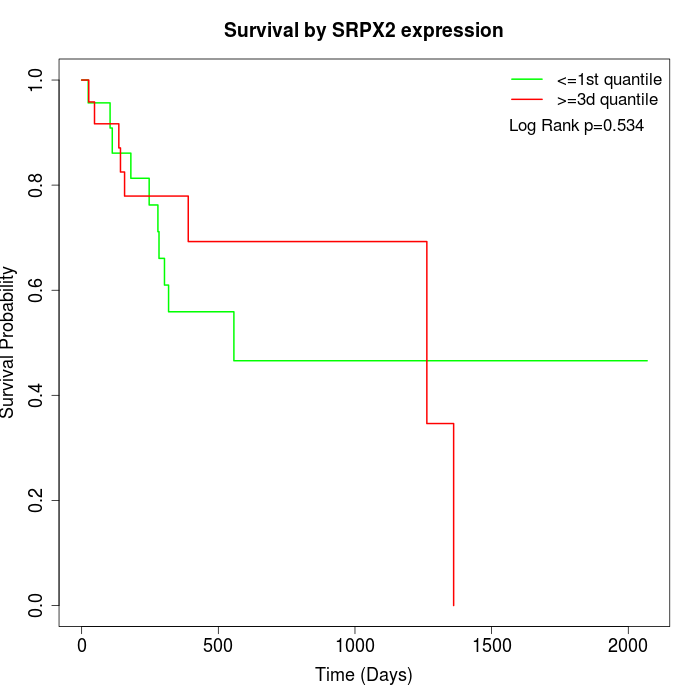
Survival by SRPX2 expression:
Note: Click image to view full size file.
Copy number change of SRPX2:
No record found for this gene.
Somatic mutations of SRPX2:
Generating mutation plots.
Highly correlated genes for SRPX2:
Showing top 20/960 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SRPX2 | SH3PXD2A-AS1 | 0.773018 | 3 | 0 | 3 |
| SRPX2 | ZNRF1 | 0.7301 | 5 | 0 | 4 |
| SRPX2 | SFTA2 | 0.715676 | 4 | 0 | 4 |
| SRPX2 | PCBP1-AS1 | 0.703008 | 3 | 0 | 3 |
| SRPX2 | SPNS2 | 0.701953 | 5 | 0 | 4 |
| SRPX2 | SH3GL1 | 0.701759 | 11 | 0 | 11 |
| SRPX2 | MIR210HG | 0.699447 | 6 | 0 | 6 |
| SRPX2 | HPSE | 0.697893 | 11 | 0 | 11 |
| SRPX2 | TMEM154 | 0.694525 | 6 | 0 | 6 |
| SRPX2 | ASCC2 | 0.694492 | 11 | 0 | 10 |
| SRPX2 | PLIN3 | 0.690976 | 12 | 0 | 12 |
| SRPX2 | TOM1 | 0.690768 | 11 | 0 | 11 |
| SRPX2 | SYTL5 | 0.687105 | 3 | 0 | 3 |
| SRPX2 | PDLIM2 | 0.684655 | 10 | 0 | 10 |
| SRPX2 | MXD1 | 0.681167 | 10 | 0 | 10 |
| SRPX2 | SLC12A6 | 0.680534 | 6 | 0 | 6 |
| SRPX2 | TSPAN6 | 0.675097 | 11 | 0 | 10 |
| SRPX2 | CHMP5 | 0.674818 | 5 | 0 | 5 |
| SRPX2 | ESPL1 | 0.674808 | 10 | 0 | 8 |
| SRPX2 | TJP3 | 0.674143 | 9 | 0 | 8 |
For details and further investigation, click here