| Full name: tudor domain containing 10 | Alias Symbol: DKFZp434M202 | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 126668 | HGNC ID: HGNC:25316 | Ensembl Gene: ENSG00000163239 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TDRD10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TDRD10 | 126668 | 231371_at | -0.1766 | 0.4277 | |
| GSE26886 | TDRD10 | 126668 | 231371_at | -0.1474 | 0.2551 | |
| GSE45670 | TDRD10 | 126668 | 231371_at | -0.4128 | 0.0031 | |
| GSE53622 | TDRD10 | 126668 | 77419 | -0.2117 | 0.0594 | |
| GSE53624 | TDRD10 | 126668 | 97073 | -1.4578 | 0.0000 | |
| GSE63941 | TDRD10 | 126668 | 231371_at | 0.2947 | 0.1027 | |
| GSE77861 | TDRD10 | 126668 | 231371_at | 0.0539 | 0.5643 | |
| GSE97050 | TDRD10 | 126668 | A_33_P3267250 | 0.0036 | 0.9882 | |
| SRP064894 | TDRD10 | 126668 | RNAseq | -0.1224 | 0.7575 | |
| SRP133303 | TDRD10 | 126668 | RNAseq | -1.1845 | 0.0011 | |
| SRP219564 | TDRD10 | 126668 | RNAseq | 1.4306 | 0.1292 | |
| TCGA | TDRD10 | 126668 | RNAseq | -1.0105 | 0.0492 |
Upregulated datasets: 0; Downregulated datasets: 3.
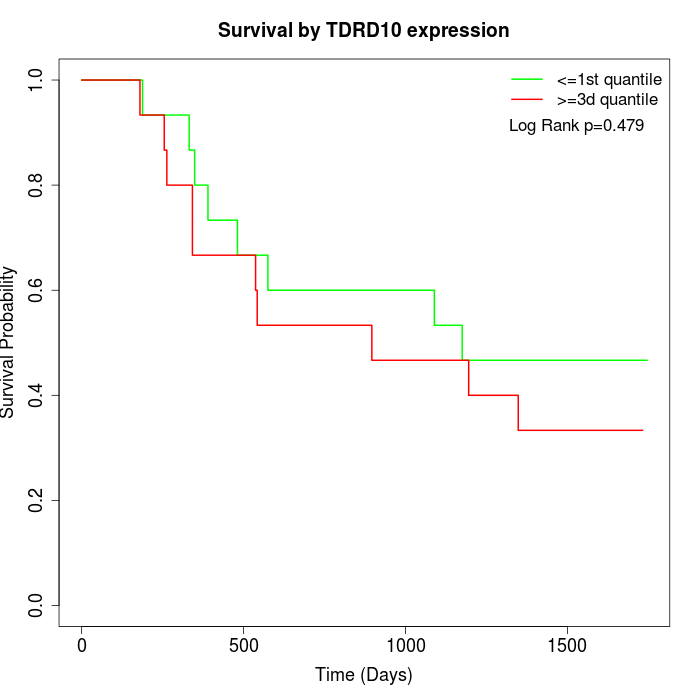
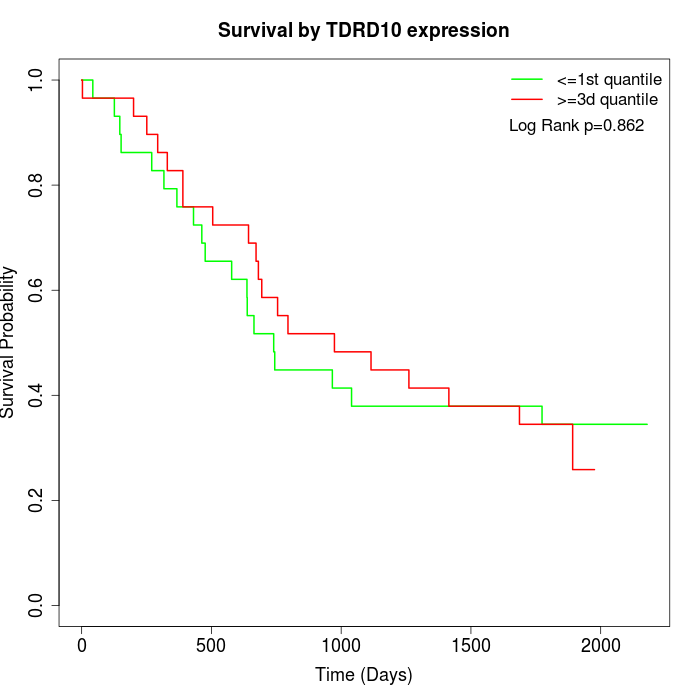
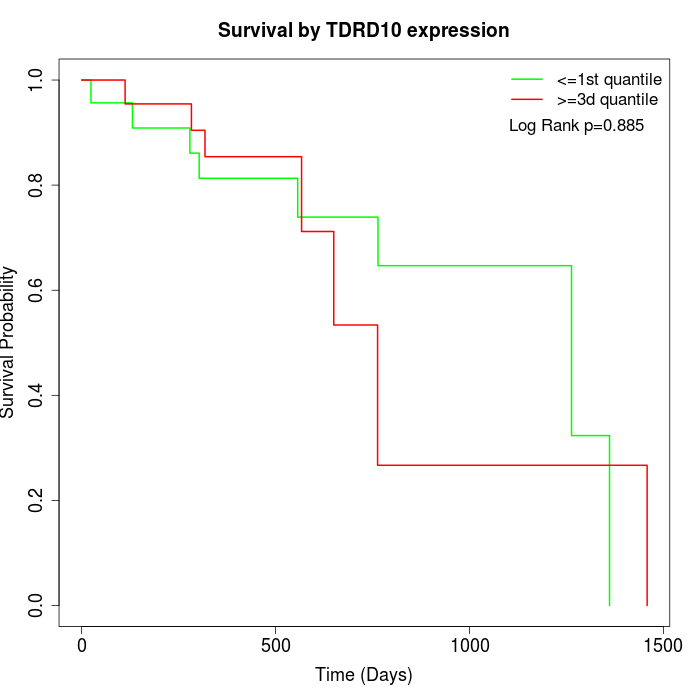
Survival by TDRD10 expression:
Note: Click image to view full size file.
Copy number change of TDRD10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TDRD10 | 126668 | 15 | 0 | 15 | |
| GSE20123 | TDRD10 | 126668 | 14 | 0 | 16 | |
| GSE43470 | TDRD10 | 126668 | 6 | 2 | 35 | |
| GSE46452 | TDRD10 | 126668 | 2 | 1 | 56 | |
| GSE47630 | TDRD10 | 126668 | 14 | 0 | 26 | |
| GSE54993 | TDRD10 | 126668 | 0 | 4 | 66 | |
| GSE54994 | TDRD10 | 126668 | 16 | 0 | 37 | |
| GSE60625 | TDRD10 | 126668 | 0 | 0 | 11 | |
| GSE74703 | TDRD10 | 126668 | 6 | 1 | 29 | |
| GSE74704 | TDRD10 | 126668 | 7 | 0 | 13 | |
| TCGA | TDRD10 | 126668 | 38 | 2 | 56 |
Total number of gains: 118; Total number of losses: 10; Total Number of normals: 360.
Somatic mutations of TDRD10:
Generating mutation plots.
Highly correlated genes for TDRD10:
Showing top 20/48 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TDRD10 | TNXB | 0.784596 | 3 | 0 | 3 |
| TDRD10 | ZMYND10 | 0.739598 | 3 | 0 | 3 |
| TDRD10 | GATA2 | 0.73022 | 3 | 0 | 3 |
| TDRD10 | FUZ | 0.728322 | 3 | 0 | 3 |
| TDRD10 | GPAT2 | 0.683504 | 3 | 0 | 3 |
| TDRD10 | FGD5 | 0.673321 | 4 | 0 | 4 |
| TDRD10 | VWDE | 0.660922 | 3 | 0 | 3 |
| TDRD10 | DGKZ | 0.659591 | 3 | 0 | 3 |
| TDRD10 | TIGD5 | 0.641497 | 3 | 0 | 3 |
| TDRD10 | FGF14 | 0.634688 | 3 | 0 | 3 |
| TDRD10 | WAS | 0.631061 | 3 | 0 | 3 |
| TDRD10 | NXPH3 | 0.623685 | 4 | 0 | 3 |
| TDRD10 | NRG2 | 0.623084 | 4 | 0 | 3 |
| TDRD10 | RAB17 | 0.619501 | 3 | 0 | 3 |
| TDRD10 | PDZD2 | 0.61305 | 5 | 0 | 4 |
| TDRD10 | CTSG | 0.611569 | 4 | 0 | 3 |
| TDRD10 | SFRP1 | 0.609249 | 4 | 0 | 3 |
| TDRD10 | AGPAT2 | 0.607896 | 4 | 0 | 3 |
| TDRD10 | SHE | 0.607539 | 5 | 0 | 4 |
| TDRD10 | KCNC3 | 0.60546 | 4 | 0 | 3 |
For details and further investigation, click here