| Full name: TNF receptor superfamily member 10b | Alias Symbol: DR5|KILLER|TRICK2A|TRAIL-R2|TRICKB|CD262|TRAILR2 | ||
| Type: protein-coding gene | Cytoband: 8p21.3 | ||
| Entrez ID: 8795 | HGNC ID: HGNC:11905 | Ensembl Gene: ENSG00000120889 | OMIM ID: 603612 |
| Drug and gene relationship at DGIdb | |||
TNFRSF10B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04115 | p53 signaling pathway | |
| hsa04210 | Apoptosis | |
| hsa04650 | Natural killer cell mediated cytotoxicity | |
| hsa05162 | Measles |
Expression of TNFRSF10B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNFRSF10B | 8795 | 209295_at | 1.1938 | 0.1310 | |
| GSE20347 | TNFRSF10B | 8795 | 209295_at | 1.1664 | 0.0000 | |
| GSE23400 | TNFRSF10B | 8795 | 209295_at | 0.9432 | 0.0000 | |
| GSE26886 | TNFRSF10B | 8795 | 209295_at | 2.1505 | 0.0000 | |
| GSE29001 | TNFRSF10B | 8795 | 209295_at | 0.8383 | 0.0033 | |
| GSE38129 | TNFRSF10B | 8795 | 209295_at | 0.9571 | 0.0000 | |
| GSE45670 | TNFRSF10B | 8795 | 209295_at | 0.7582 | 0.0061 | |
| GSE53622 | TNFRSF10B | 8795 | 157904 | 1.0347 | 0.0000 | |
| GSE53624 | TNFRSF10B | 8795 | 157904 | 1.0679 | 0.0000 | |
| GSE63941 | TNFRSF10B | 8795 | 209295_at | -1.5961 | 0.0082 | |
| GSE77861 | TNFRSF10B | 8795 | 209295_at | 1.2454 | 0.0605 | |
| GSE97050 | TNFRSF10B | 8795 | A_24_P218265 | 0.8502 | 0.0665 | |
| SRP007169 | TNFRSF10B | 8795 | RNAseq | 2.5549 | 0.0000 | |
| SRP008496 | TNFRSF10B | 8795 | RNAseq | 2.6484 | 0.0000 | |
| SRP064894 | TNFRSF10B | 8795 | RNAseq | 0.8694 | 0.0024 | |
| SRP133303 | TNFRSF10B | 8795 | RNAseq | 0.8267 | 0.0009 | |
| SRP159526 | TNFRSF10B | 8795 | RNAseq | 0.4497 | 0.3225 | |
| SRP193095 | TNFRSF10B | 8795 | RNAseq | 1.1847 | 0.0000 | |
| SRP219564 | TNFRSF10B | 8795 | RNAseq | 0.5270 | 0.0905 | |
| TCGA | TNFRSF10B | 8795 | RNAseq | 0.1879 | 0.0085 |
Upregulated datasets: 7; Downregulated datasets: 1.
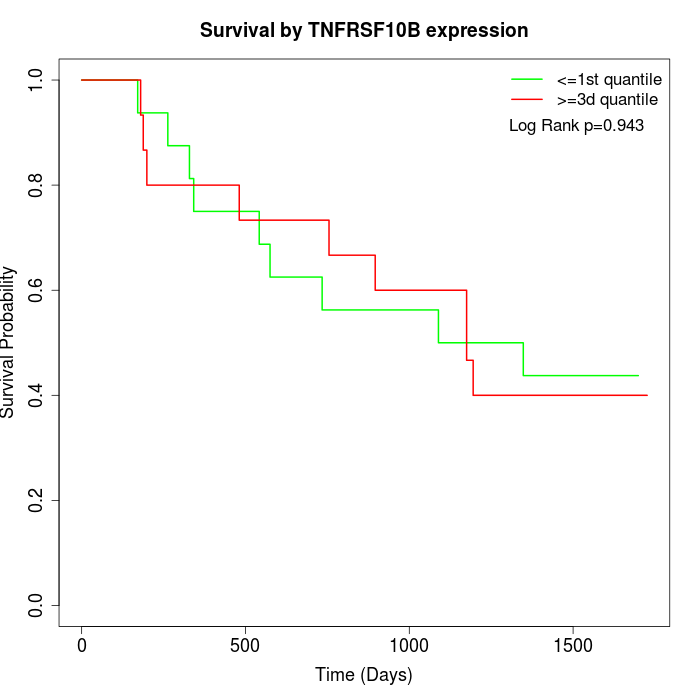
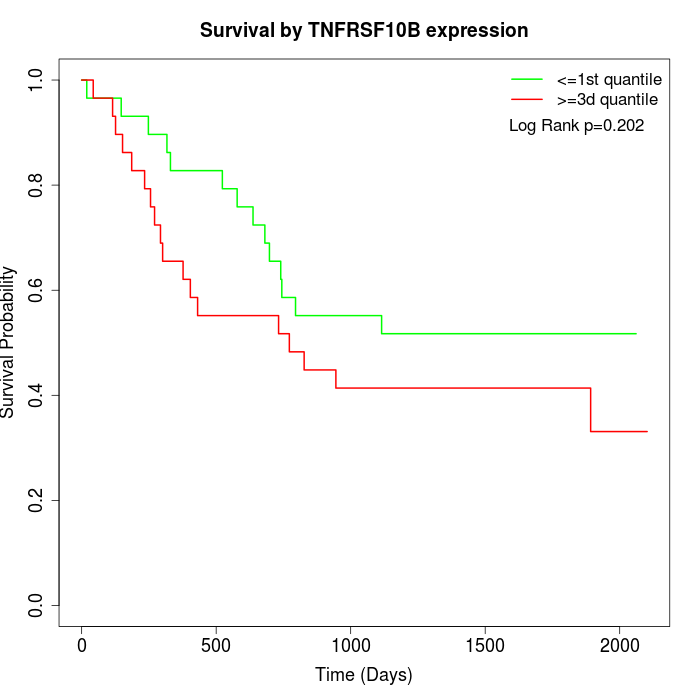
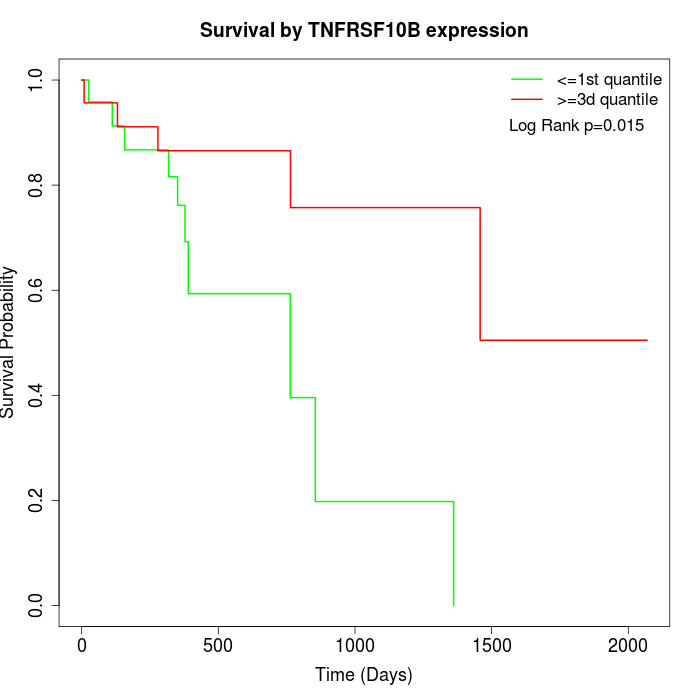
Survival by TNFRSF10B expression:
Note: Click image to view full size file.
Copy number change of TNFRSF10B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNFRSF10B | 8795 | 4 | 11 | 15 | |
| GSE20123 | TNFRSF10B | 8795 | 4 | 11 | 15 | |
| GSE43470 | TNFRSF10B | 8795 | 4 | 9 | 30 | |
| GSE46452 | TNFRSF10B | 8795 | 13 | 13 | 33 | |
| GSE47630 | TNFRSF10B | 8795 | 10 | 8 | 22 | |
| GSE54993 | TNFRSF10B | 8795 | 2 | 14 | 54 | |
| GSE54994 | TNFRSF10B | 8795 | 8 | 18 | 27 | |
| GSE60625 | TNFRSF10B | 8795 | 3 | 0 | 8 | |
| GSE74703 | TNFRSF10B | 8795 | 4 | 7 | 25 | |
| GSE74704 | TNFRSF10B | 8795 | 3 | 8 | 9 | |
| TCGA | TNFRSF10B | 8795 | 14 | 43 | 39 |
Total number of gains: 69; Total number of losses: 142; Total Number of normals: 277.
Somatic mutations of TNFRSF10B:
Generating mutation plots.
Highly correlated genes for TNFRSF10B:
Showing top 20/1407 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNFRSF10B | RICTOR | 0.807343 | 3 | 0 | 3 |
| TNFRSF10B | LRCH3 | 0.792703 | 4 | 0 | 3 |
| TNFRSF10B | CPT1C | 0.788427 | 3 | 0 | 3 |
| TNFRSF10B | CERS5 | 0.772464 | 3 | 0 | 3 |
| TNFRSF10B | DOCK11 | 0.771014 | 3 | 0 | 3 |
| TNFRSF10B | CYTH3 | 0.765974 | 3 | 0 | 3 |
| TNFRSF10B | UTP23 | 0.760248 | 3 | 0 | 3 |
| TNFRSF10B | TMEM181 | 0.75984 | 3 | 0 | 3 |
| TNFRSF10B | RRM2B | 0.754066 | 4 | 0 | 4 |
| TNFRSF10B | PDE7A | 0.749871 | 6 | 0 | 6 |
| TNFRSF10B | ZNF469 | 0.746559 | 5 | 0 | 5 |
| TNFRSF10B | WDR81 | 0.742513 | 3 | 0 | 3 |
| TNFRSF10B | MEX3B | 0.742389 | 3 | 0 | 3 |
| TNFRSF10B | PRTFDC1 | 0.728784 | 4 | 0 | 4 |
| TNFRSF10B | MET | 0.724046 | 9 | 0 | 9 |
| TNFRSF10B | VSTM4 | 0.722536 | 3 | 0 | 3 |
| TNFRSF10B | SLC39A10 | 0.721778 | 5 | 0 | 5 |
| TNFRSF10B | PAPPA | 0.719842 | 3 | 0 | 3 |
| TNFRSF10B | COPB1 | 0.718767 | 5 | 0 | 4 |
| TNFRSF10B | COQ10B | 0.718422 | 3 | 0 | 3 |
For details and further investigation, click here