| Full name: tripartite motif containing 14 | Alias Symbol: KIAA0129 | ||
| Type: protein-coding gene | Cytoband: 9q22.33 | ||
| Entrez ID: 9830 | HGNC ID: HGNC:16283 | Ensembl Gene: ENSG00000106785 | OMIM ID: 606556 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM14 | 9830 | 203148_s_at | -0.2406 | 0.7580 | |
| GSE20347 | TRIM14 | 9830 | 203148_s_at | -0.5737 | 0.0092 | |
| GSE23400 | TRIM14 | 9830 | 203147_s_at | -0.1348 | 0.0164 | |
| GSE26886 | TRIM14 | 9830 | 203148_s_at | -0.6317 | 0.0040 | |
| GSE29001 | TRIM14 | 9830 | 203148_s_at | -0.1640 | 0.6344 | |
| GSE38129 | TRIM14 | 9830 | 203148_s_at | -0.0632 | 0.8023 | |
| GSE45670 | TRIM14 | 9830 | 203148_s_at | 0.2018 | 0.4602 | |
| GSE53622 | TRIM14 | 9830 | 51770 | 0.0895 | 0.3372 | |
| GSE53624 | TRIM14 | 9830 | 16375 | -0.0811 | 0.2496 | |
| GSE63941 | TRIM14 | 9830 | 203148_s_at | 0.3933 | 0.5647 | |
| GSE77861 | TRIM14 | 9830 | 203148_s_at | -0.5344 | 0.0298 | |
| GSE97050 | TRIM14 | 9830 | A_23_P425752 | 0.6953 | 0.2621 | |
| SRP007169 | TRIM14 | 9830 | RNAseq | 0.2683 | 0.5511 | |
| SRP008496 | TRIM14 | 9830 | RNAseq | -0.2786 | 0.4070 | |
| SRP064894 | TRIM14 | 9830 | RNAseq | 0.1295 | 0.5370 | |
| SRP133303 | TRIM14 | 9830 | RNAseq | -0.3630 | 0.1291 | |
| SRP159526 | TRIM14 | 9830 | RNAseq | -0.8448 | 0.0426 | |
| SRP193095 | TRIM14 | 9830 | RNAseq | -0.1604 | 0.3279 | |
| SRP219564 | TRIM14 | 9830 | RNAseq | -0.2865 | 0.5262 | |
| TCGA | TRIM14 | 9830 | RNAseq | 0.2627 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 0.
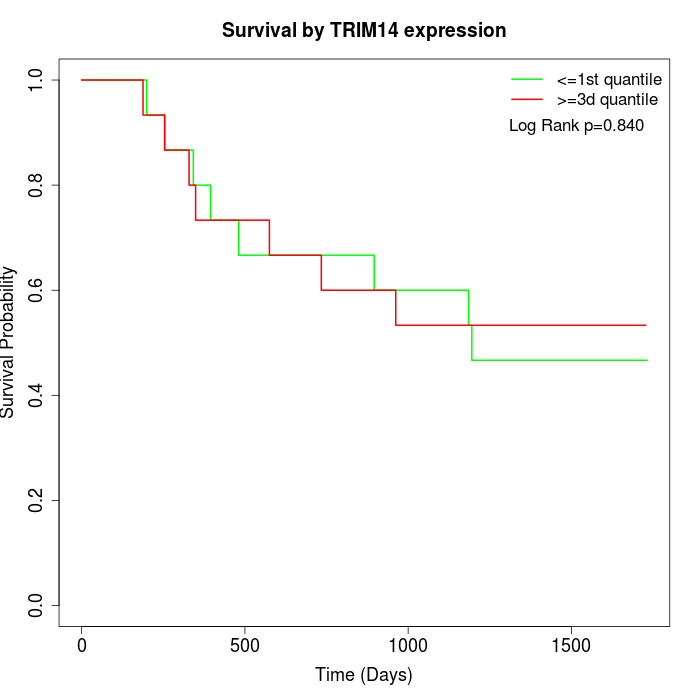
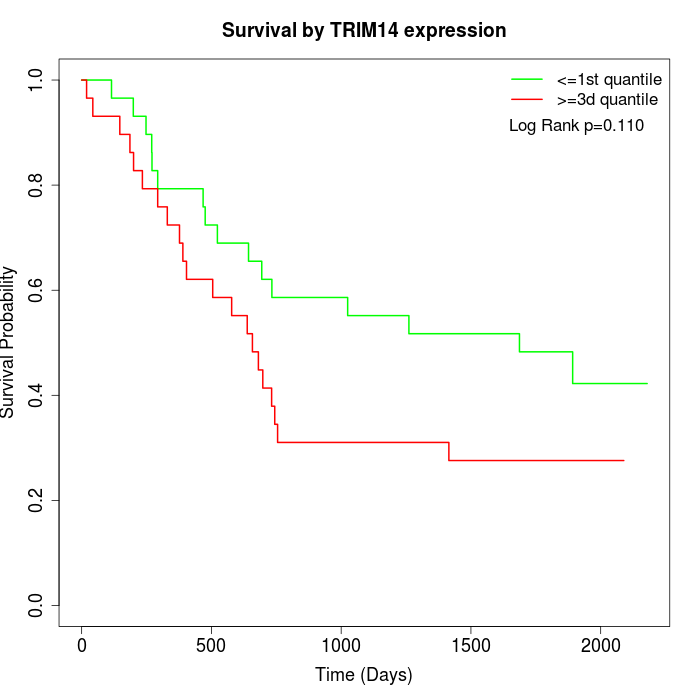
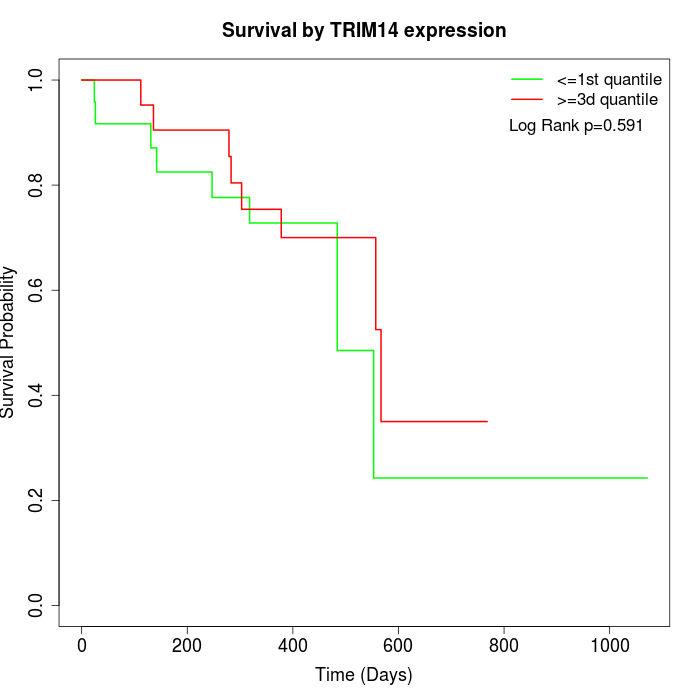
Survival by TRIM14 expression:
Note: Click image to view full size file.
Copy number change of TRIM14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM14 | 9830 | 5 | 8 | 17 | |
| GSE20123 | TRIM14 | 9830 | 6 | 7 | 17 | |
| GSE43470 | TRIM14 | 9830 | 6 | 3 | 34 | |
| GSE46452 | TRIM14 | 9830 | 6 | 14 | 39 | |
| GSE47630 | TRIM14 | 9830 | 1 | 17 | 22 | |
| GSE54993 | TRIM14 | 9830 | 3 | 3 | 64 | |
| GSE54994 | TRIM14 | 9830 | 8 | 11 | 34 | |
| GSE60625 | TRIM14 | 9830 | 0 | 0 | 11 | |
| GSE74703 | TRIM14 | 9830 | 5 | 3 | 28 | |
| GSE74704 | TRIM14 | 9830 | 3 | 5 | 12 | |
| TCGA | TRIM14 | 9830 | 27 | 22 | 47 |
Total number of gains: 70; Total number of losses: 93; Total Number of normals: 325.
Somatic mutations of TRIM14:
Generating mutation plots.
Highly correlated genes for TRIM14:
Showing top 20/200 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM14 | CARD6 | 0.73325 | 3 | 0 | 3 |
| TRIM14 | ARHGAP27 | 0.723157 | 3 | 0 | 3 |
| TRIM14 | GGT6 | 0.666532 | 4 | 0 | 4 |
| TRIM14 | FAM3B | 0.664597 | 3 | 0 | 3 |
| TRIM14 | SH2D1B | 0.652985 | 4 | 0 | 4 |
| TRIM14 | GPT2 | 0.650545 | 3 | 0 | 3 |
| TRIM14 | TMPRSS2 | 0.644643 | 4 | 0 | 4 |
| TRIM14 | VAMP8 | 0.643537 | 7 | 0 | 6 |
| TRIM14 | C15orf48 | 0.642074 | 4 | 0 | 4 |
| TRIM14 | KLB | 0.640026 | 3 | 0 | 3 |
| TRIM14 | SPNS2 | 0.634952 | 3 | 0 | 3 |
| TRIM14 | HERC6 | 0.630803 | 10 | 0 | 9 |
| TRIM14 | ZNF519 | 0.625199 | 3 | 0 | 3 |
| TRIM14 | WDFY1 | 0.622654 | 4 | 0 | 4 |
| TRIM14 | TMEM144 | 0.621447 | 5 | 0 | 4 |
| TRIM14 | MAL2 | 0.620477 | 5 | 0 | 4 |
| TRIM14 | ATP13A4 | 0.615674 | 3 | 0 | 3 |
| TRIM14 | PRSS8 | 0.613895 | 5 | 0 | 3 |
| TRIM14 | BBOX1 | 0.613182 | 5 | 0 | 5 |
| TRIM14 | FAM3D | 0.608921 | 3 | 0 | 3 |
For details and further investigation, click here