| Full name: tripartite motif containing 62 | Alias Symbol: FLJ10759|DEAR1 | ||
| Type: protein-coding gene | Cytoband: 1p35.1 | ||
| Entrez ID: 55223 | HGNC ID: HGNC:25574 | Ensembl Gene: ENSG00000116525 | OMIM ID: 616755 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM62:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM62 | 55223 | 219272_at | 0.2315 | 0.4399 | |
| GSE20347 | TRIM62 | 55223 | 58308_at | -0.1017 | 0.2695 | |
| GSE23400 | TRIM62 | 55223 | 219272_at | -0.1865 | 0.0001 | |
| GSE26886 | TRIM62 | 55223 | 58308_at | -0.0264 | 0.8830 | |
| GSE29001 | TRIM62 | 55223 | 58308_at | -0.0408 | 0.8775 | |
| GSE38129 | TRIM62 | 55223 | 58308_at | -0.1134 | 0.3380 | |
| GSE45670 | TRIM62 | 55223 | 58308_at | 0.1514 | 0.2357 | |
| GSE53622 | TRIM62 | 55223 | 15904 | 0.1982 | 0.0668 | |
| GSE53624 | TRIM62 | 55223 | 15904 | 0.3153 | 0.0003 | |
| GSE63941 | TRIM62 | 55223 | 58308_at | -0.0740 | 0.8215 | |
| GSE77861 | TRIM62 | 55223 | 219272_at | -0.0748 | 0.6031 | |
| GSE97050 | TRIM62 | 55223 | A_33_P3315423 | 0.3671 | 0.3308 | |
| SRP007169 | TRIM62 | 55223 | RNAseq | -0.5607 | 0.1993 | |
| SRP064894 | TRIM62 | 55223 | RNAseq | -0.0298 | 0.8682 | |
| SRP133303 | TRIM62 | 55223 | RNAseq | -0.0193 | 0.9132 | |
| SRP159526 | TRIM62 | 55223 | RNAseq | -0.4736 | 0.0799 | |
| SRP193095 | TRIM62 | 55223 | RNAseq | 0.0916 | 0.4596 | |
| SRP219564 | TRIM62 | 55223 | RNAseq | -0.2944 | 0.4037 | |
| TCGA | TRIM62 | 55223 | RNAseq | 0.0856 | 0.1728 |
Upregulated datasets: 0; Downregulated datasets: 0.
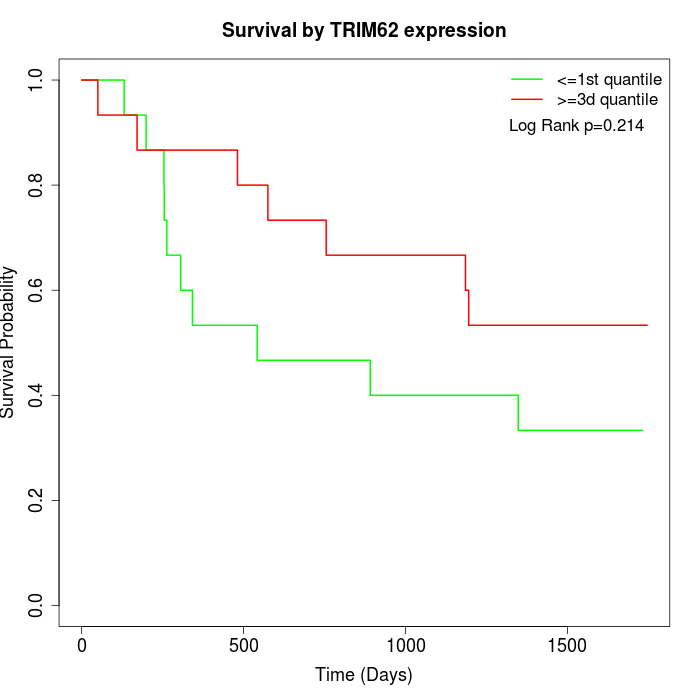
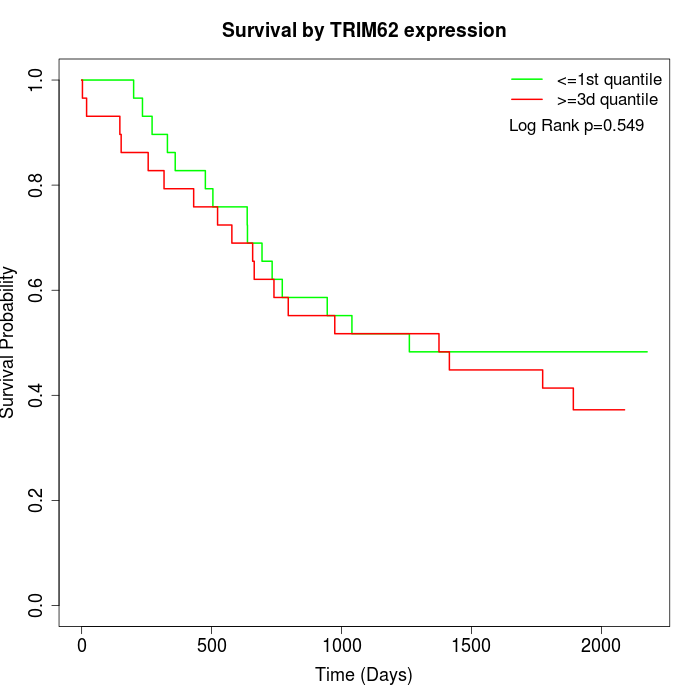
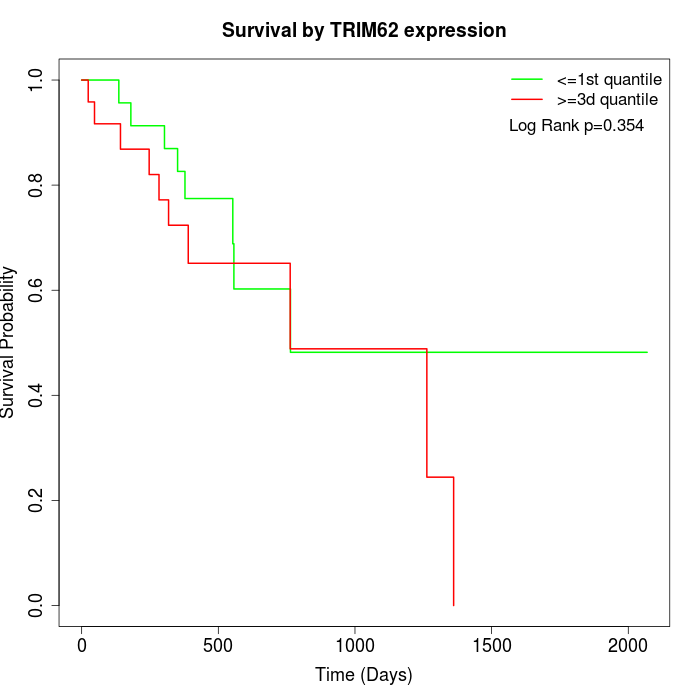
Survival by TRIM62 expression:
Note: Click image to view full size file.
Copy number change of TRIM62:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM62 | 55223 | 2 | 5 | 23 | |
| GSE20123 | TRIM62 | 55223 | 2 | 4 | 24 | |
| GSE43470 | TRIM62 | 55223 | 5 | 4 | 34 | |
| GSE46452 | TRIM62 | 55223 | 4 | 1 | 54 | |
| GSE47630 | TRIM62 | 55223 | 8 | 3 | 29 | |
| GSE54993 | TRIM62 | 55223 | 1 | 1 | 68 | |
| GSE54994 | TRIM62 | 55223 | 11 | 2 | 40 | |
| GSE60625 | TRIM62 | 55223 | 0 | 0 | 11 | |
| GSE74703 | TRIM62 | 55223 | 4 | 3 | 29 | |
| GSE74704 | TRIM62 | 55223 | 1 | 0 | 19 | |
| TCGA | TRIM62 | 55223 | 13 | 18 | 65 |
Total number of gains: 51; Total number of losses: 41; Total Number of normals: 396.
Somatic mutations of TRIM62:
Generating mutation plots.
Highly correlated genes for TRIM62:
Showing top 20/484 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM62 | DERL3 | 0.758325 | 3 | 0 | 3 |
| TRIM62 | GET4 | 0.722934 | 3 | 0 | 3 |
| TRIM62 | MMS19 | 0.721408 | 3 | 0 | 3 |
| TRIM62 | CPT1C | 0.721068 | 3 | 0 | 3 |
| TRIM62 | HPS4 | 0.714813 | 3 | 0 | 3 |
| TRIM62 | ZFC3H1 | 0.706676 | 3 | 0 | 3 |
| TRIM62 | DVL1 | 0.704593 | 4 | 0 | 4 |
| TRIM62 | TOP3A | 0.702283 | 5 | 0 | 5 |
| TRIM62 | RRAGC | 0.696127 | 3 | 0 | 3 |
| TRIM62 | LZTS1 | 0.693115 | 5 | 0 | 4 |
| TRIM62 | SARDH | 0.693047 | 4 | 0 | 4 |
| TRIM62 | TP53I11 | 0.689964 | 3 | 0 | 3 |
| TRIM62 | ISY1 | 0.688623 | 3 | 0 | 3 |
| TRIM62 | MAPK11 | 0.686141 | 4 | 0 | 3 |
| TRIM62 | ATAT1 | 0.686118 | 3 | 0 | 3 |
| TRIM62 | IER3IP1 | 0.669058 | 3 | 0 | 3 |
| TRIM62 | POU2F2 | 0.66602 | 6 | 0 | 5 |
| TRIM62 | HERC4 | 0.665992 | 3 | 0 | 3 |
| TRIM62 | TAF1C | 0.665748 | 4 | 0 | 3 |
| TRIM62 | GNL3L | 0.665147 | 3 | 0 | 3 |
For details and further investigation, click here