| Full name: villin 1 | Alias Symbol: D2S1471 | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 7429 | HGNC ID: HGNC:12690 | Ensembl Gene: ENSG00000127831 | OMIM ID: 193040 |
| Drug and gene relationship at DGIdb | |||
Expression of VIL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VIL1 | 7429 | 1554943_at | -0.0270 | 0.9431 | |
| GSE20347 | VIL1 | 7429 | 205506_at | 0.0152 | 0.8889 | |
| GSE23400 | VIL1 | 7429 | 205506_at | -0.1612 | 0.0000 | |
| GSE26886 | VIL1 | 7429 | 228912_at | 0.0527 | 0.6365 | |
| GSE29001 | VIL1 | 7429 | 205506_at | -0.1469 | 0.2753 | |
| GSE38129 | VIL1 | 7429 | 205506_at | -0.0215 | 0.8602 | |
| GSE45670 | VIL1 | 7429 | 1554943_at | 0.0449 | 0.5199 | |
| GSE53622 | VIL1 | 7429 | 40635 | 0.5001 | 0.0137 | |
| GSE53624 | VIL1 | 7429 | 40635 | 0.7172 | 0.0001 | |
| GSE63941 | VIL1 | 7429 | 1554943_at | 0.0323 | 0.8268 | |
| GSE77861 | VIL1 | 7429 | 205506_at | -0.0250 | 0.8651 | |
| SRP133303 | VIL1 | 7429 | RNAseq | -0.4180 | 0.1431 | |
| TCGA | VIL1 | 7429 | RNAseq | -1.6276 | 0.0528 |
Upregulated datasets: 0; Downregulated datasets: 0.
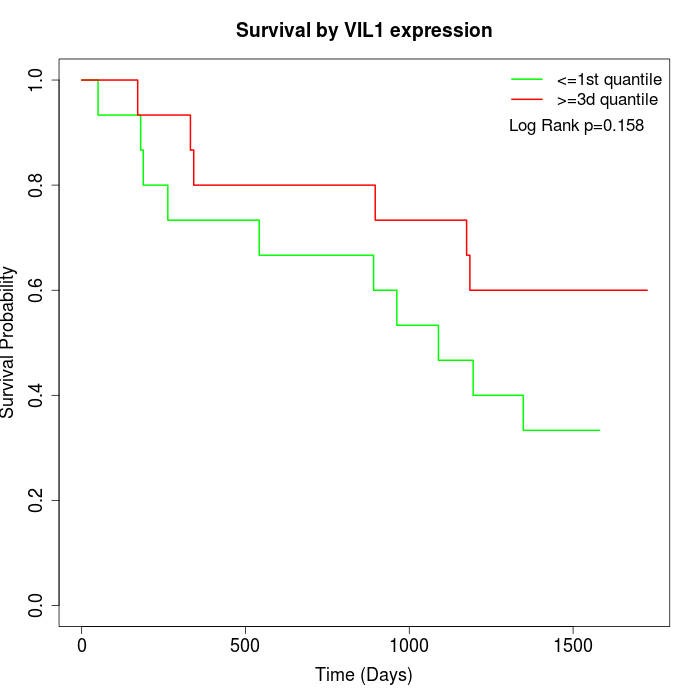
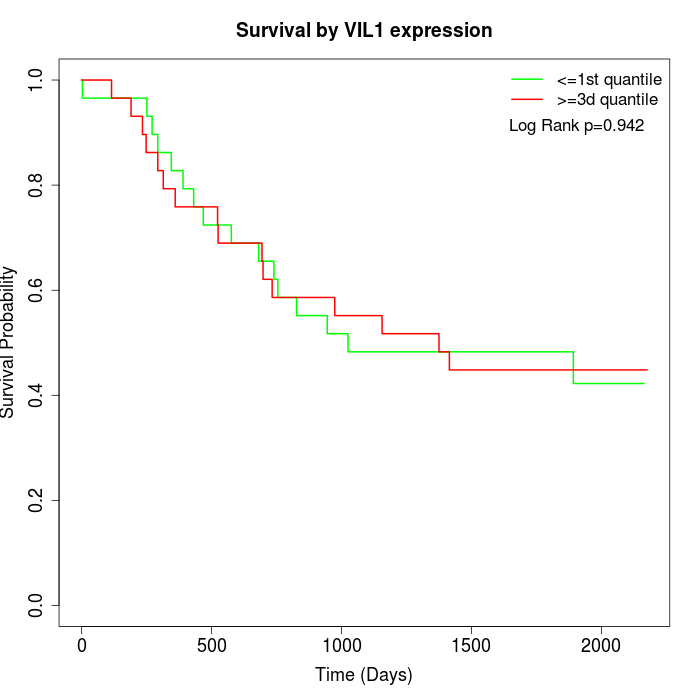
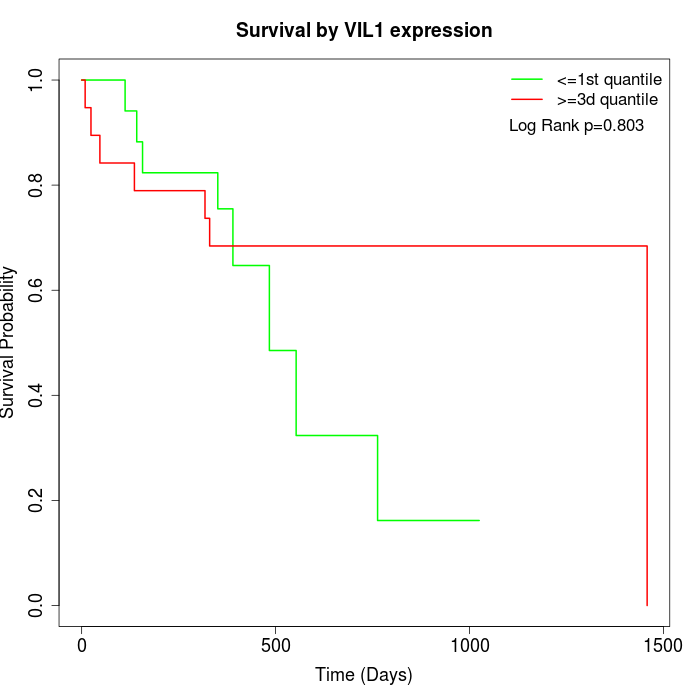
Survival by VIL1 expression:
Note: Click image to view full size file.
Copy number change of VIL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VIL1 | 7429 | 1 | 14 | 15 | |
| GSE20123 | VIL1 | 7429 | 1 | 13 | 16 | |
| GSE43470 | VIL1 | 7429 | 1 | 7 | 35 | |
| GSE46452 | VIL1 | 7429 | 0 | 5 | 54 | |
| GSE47630 | VIL1 | 7429 | 4 | 5 | 31 | |
| GSE54993 | VIL1 | 7429 | 1 | 2 | 67 | |
| GSE54994 | VIL1 | 7429 | 8 | 10 | 35 | |
| GSE60625 | VIL1 | 7429 | 0 | 3 | 8 | |
| GSE74703 | VIL1 | 7429 | 1 | 5 | 30 | |
| GSE74704 | VIL1 | 7429 | 1 | 7 | 12 | |
| TCGA | VIL1 | 7429 | 11 | 29 | 56 |
Total number of gains: 29; Total number of losses: 100; Total Number of normals: 359.
Somatic mutations of VIL1:
Generating mutation plots.
Highly correlated genes for VIL1:
Showing top 20/297 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VIL1 | RAX | 0.692884 | 3 | 0 | 3 |
| VIL1 | PRKG1 | 0.682254 | 3 | 0 | 3 |
| VIL1 | LOXL3 | 0.657773 | 3 | 0 | 3 |
| VIL1 | OR3A1 | 0.648767 | 3 | 0 | 3 |
| VIL1 | TCAP | 0.645232 | 3 | 0 | 3 |
| VIL1 | GABRA5 | 0.640975 | 3 | 0 | 3 |
| VIL1 | BRINP2 | 0.640236 | 5 | 0 | 5 |
| VIL1 | GRIN1 | 0.627606 | 4 | 0 | 3 |
| VIL1 | ARRB1 | 0.624075 | 3 | 0 | 3 |
| VIL1 | MT3 | 0.622351 | 4 | 0 | 4 |
| VIL1 | HTR3A | 0.618397 | 5 | 0 | 5 |
| VIL1 | OCM2 | 0.618059 | 3 | 0 | 3 |
| VIL1 | SIGLEC5 | 0.614407 | 4 | 0 | 4 |
| VIL1 | MASP1 | 0.614042 | 3 | 0 | 3 |
| VIL1 | PRPH | 0.611769 | 4 | 0 | 3 |
| VIL1 | PON1 | 0.610138 | 4 | 0 | 3 |
| VIL1 | BCAN | 0.609252 | 6 | 0 | 5 |
| VIL1 | FGA | 0.60875 | 4 | 0 | 3 |
| VIL1 | CNGB1 | 0.608327 | 4 | 0 | 3 |
| VIL1 | EBF2 | 0.608208 | 3 | 0 | 3 |
For details and further investigation, click here