| Full name: acid phosphatase 2, lysosomal | Alias Symbol: LAP | ||
| Type: protein-coding gene | Cytoband: 11p11.2 | ||
| Entrez ID: 53 | HGNC ID: HGNC:123 | Ensembl Gene: ENSG00000134575 | OMIM ID: 171650 |
| Drug and gene relationship at DGIdb | |||
Expression of ACP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACP2 | 53 | 202767_at | 0.3263 | 0.3997 | |
| GSE20347 | ACP2 | 53 | 202767_at | 0.2172 | 0.0560 | |
| GSE23400 | ACP2 | 53 | 202767_at | 0.3667 | 0.0000 | |
| GSE26886 | ACP2 | 53 | 202767_at | 0.0995 | 0.6352 | |
| GSE29001 | ACP2 | 53 | 202767_at | 0.5291 | 0.0030 | |
| GSE38129 | ACP2 | 53 | 202767_at | 0.1750 | 0.3508 | |
| GSE45670 | ACP2 | 53 | 202767_at | 0.4564 | 0.0113 | |
| GSE53622 | ACP2 | 53 | 88833 | 0.7206 | 0.0000 | |
| GSE53624 | ACP2 | 53 | 88833 | 0.6341 | 0.0000 | |
| GSE63941 | ACP2 | 53 | 202767_at | -0.6016 | 0.3371 | |
| GSE77861 | ACP2 | 53 | 202767_at | 0.1448 | 0.3846 | |
| GSE97050 | ACP2 | 53 | A_33_P3310864 | 0.9092 | 0.0966 | |
| SRP007169 | ACP2 | 53 | RNAseq | 1.1094 | 0.0723 | |
| SRP008496 | ACP2 | 53 | RNAseq | 0.7493 | 0.0652 | |
| SRP064894 | ACP2 | 53 | RNAseq | 1.2565 | 0.0000 | |
| SRP133303 | ACP2 | 53 | RNAseq | 0.5288 | 0.0004 | |
| SRP159526 | ACP2 | 53 | RNAseq | 0.6656 | 0.0132 | |
| SRP193095 | ACP2 | 53 | RNAseq | 0.2900 | 0.0956 | |
| SRP219564 | ACP2 | 53 | RNAseq | 0.8579 | 0.0105 | |
| TCGA | ACP2 | 53 | RNAseq | -0.0380 | 0.4727 |
Upregulated datasets: 1; Downregulated datasets: 0.
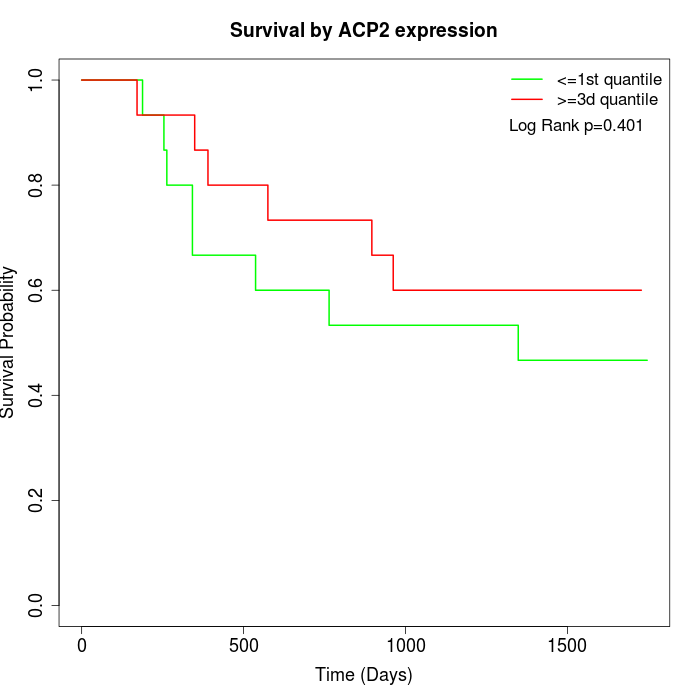
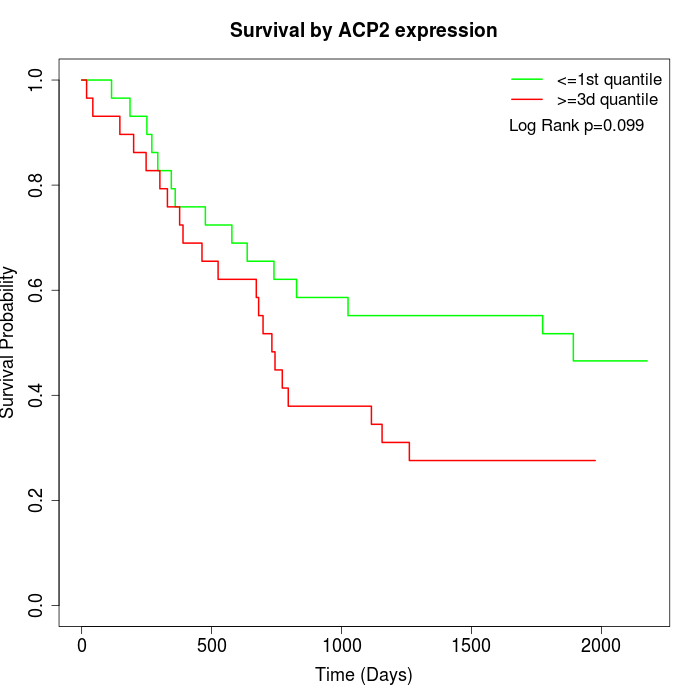
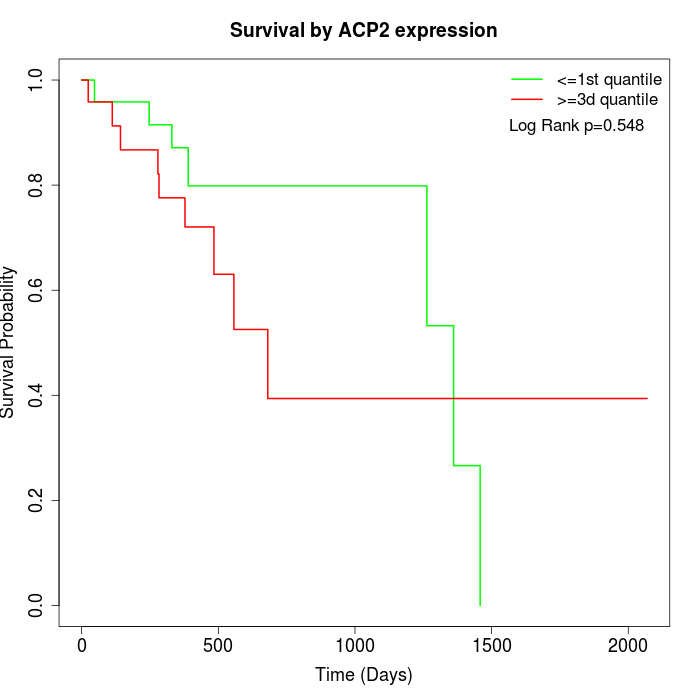
Survival by ACP2 expression:
Note: Click image to view full size file.
Copy number change of ACP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACP2 | 53 | 3 | 7 | 20 | |
| GSE20123 | ACP2 | 53 | 3 | 6 | 21 | |
| GSE43470 | ACP2 | 53 | 1 | 4 | 38 | |
| GSE46452 | ACP2 | 53 | 8 | 5 | 46 | |
| GSE47630 | ACP2 | 53 | 3 | 9 | 28 | |
| GSE54993 | ACP2 | 53 | 3 | 0 | 67 | |
| GSE54994 | ACP2 | 53 | 1 | 11 | 41 | |
| GSE60625 | ACP2 | 53 | 0 | 0 | 11 | |
| GSE74703 | ACP2 | 53 | 1 | 2 | 33 | |
| GSE74704 | ACP2 | 53 | 2 | 4 | 14 | |
| TCGA | ACP2 | 53 | 10 | 20 | 66 |
Total number of gains: 35; Total number of losses: 68; Total Number of normals: 385.
Somatic mutations of ACP2:
Generating mutation plots.
Highly correlated genes for ACP2:
Showing top 20/1081 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACP2 | PDIA5 | 0.772563 | 3 | 0 | 3 |
| ACP2 | HNRNPUL2 | 0.726298 | 3 | 0 | 3 |
| ACP2 | KTI12 | 0.722827 | 3 | 0 | 3 |
| ACP2 | SLC17A9 | 0.722754 | 3 | 0 | 3 |
| ACP2 | STEAP1B | 0.717612 | 3 | 0 | 3 |
| ACP2 | RPS19 | 0.715672 | 3 | 0 | 3 |
| ACP2 | DDX60L | 0.711544 | 4 | 0 | 3 |
| ACP2 | RNF213 | 0.709368 | 3 | 0 | 3 |
| ACP2 | LRRC8C | 0.704059 | 3 | 0 | 3 |
| ACP2 | HAVCR2 | 0.698712 | 3 | 0 | 3 |
| ACP2 | PYCR2 | 0.697927 | 3 | 0 | 3 |
| ACP2 | LRRC58 | 0.695677 | 3 | 0 | 3 |
| ACP2 | TAP1 | 0.691463 | 7 | 0 | 6 |
| ACP2 | BOLA3 | 0.690737 | 3 | 0 | 3 |
| ACP2 | ITGAE | 0.689483 | 3 | 0 | 3 |
| ACP2 | SLC35B2 | 0.687992 | 3 | 0 | 3 |
| ACP2 | ZNF281 | 0.685606 | 8 | 0 | 8 |
| ACP2 | XAB2 | 0.685386 | 3 | 0 | 3 |
| ACP2 | SNAP47 | 0.679789 | 3 | 0 | 3 |
| ACP2 | DHX57 | 0.67944 | 3 | 0 | 3 |
For details and further investigation, click here