| Full name: ADAM metallopeptidase domain 17 | Alias Symbol: cSVP|CD156B | ||
| Type: protein-coding gene | Cytoband: 2p25.1 | ||
| Entrez ID: 6868 | HGNC ID: HGNC:195 | Ensembl Gene: ENSG00000151694 | OMIM ID: 603639 |
| Drug and gene relationship at DGIdb | |||
ADAM17 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04330 | Notch signaling pathway | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ADAM17:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ADAM17 | 6868 | 213532_at | 0.8601 | 0.1193 | |
| GSE20347 | ADAM17 | 6868 | 213532_at | 0.5561 | 0.0014 | |
| GSE23400 | ADAM17 | 6868 | 213532_at | 0.6815 | 0.0000 | |
| GSE26886 | ADAM17 | 6868 | 213532_at | 1.1822 | 0.0000 | |
| GSE29001 | ADAM17 | 6868 | 205745_x_at | 0.6949 | 0.0098 | |
| GSE38129 | ADAM17 | 6868 | 213532_at | 0.8875 | 0.0000 | |
| GSE45670 | ADAM17 | 6868 | 213532_at | 0.6691 | 0.0010 | |
| GSE53622 | ADAM17 | 6868 | 44910 | 1.0723 | 0.0000 | |
| GSE53624 | ADAM17 | 6868 | 44910 | 0.7456 | 0.0000 | |
| GSE63941 | ADAM17 | 6868 | 213532_at | -0.2034 | 0.7005 | |
| GSE77861 | ADAM17 | 6868 | 213532_at | 0.6397 | 0.0081 | |
| GSE97050 | ADAM17 | 6868 | A_23_P143120 | 1.1078 | 0.0880 | |
| SRP007169 | ADAM17 | 6868 | RNAseq | 0.9701 | 0.0065 | |
| SRP008496 | ADAM17 | 6868 | RNAseq | 1.1015 | 0.0015 | |
| SRP064894 | ADAM17 | 6868 | RNAseq | 0.5168 | 0.1522 | |
| SRP133303 | ADAM17 | 6868 | RNAseq | 0.7020 | 0.0000 | |
| SRP159526 | ADAM17 | 6868 | RNAseq | 1.2256 | 0.0035 | |
| SRP193095 | ADAM17 | 6868 | RNAseq | 0.7823 | 0.0000 | |
| SRP219564 | ADAM17 | 6868 | RNAseq | 0.5001 | 0.3642 | |
| TCGA | ADAM17 | 6868 | RNAseq | 0.4383 | 0.0000 |
Upregulated datasets: 4; Downregulated datasets: 0.
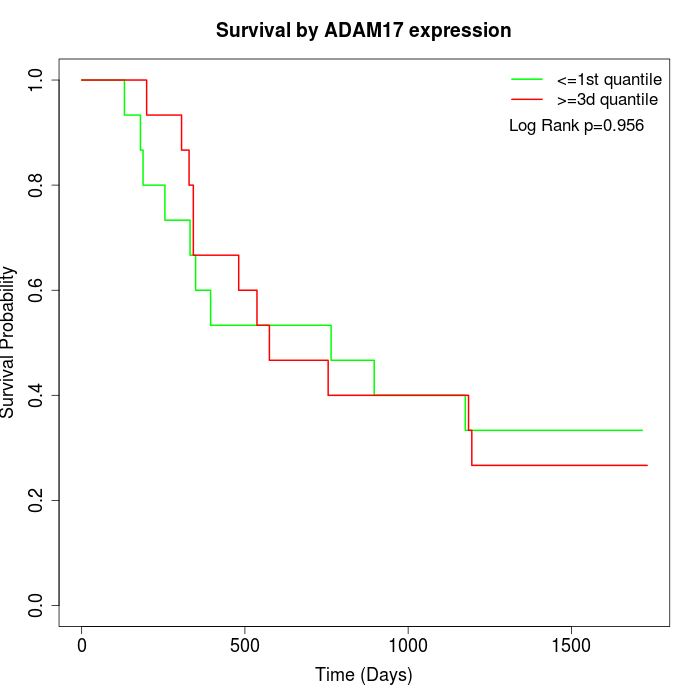
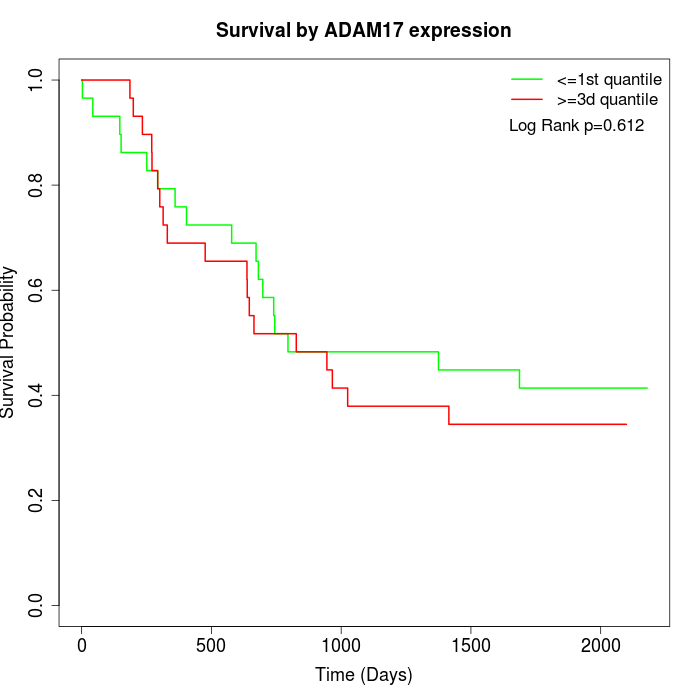
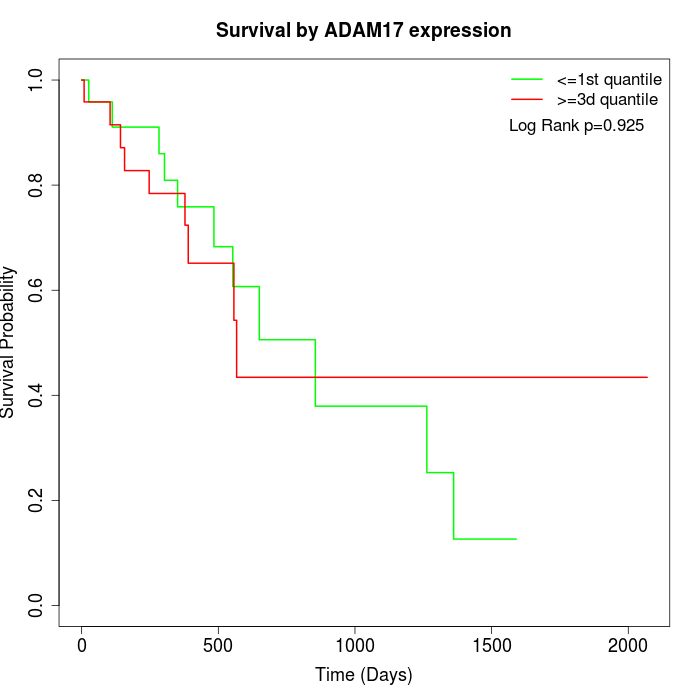
Survival by ADAM17 expression:
Note: Click image to view full size file.
Copy number change of ADAM17:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ADAM17 | 6868 | 11 | 2 | 17 | |
| GSE20123 | ADAM17 | 6868 | 11 | 2 | 17 | |
| GSE43470 | ADAM17 | 6868 | 2 | 1 | 40 | |
| GSE46452 | ADAM17 | 6868 | 3 | 4 | 52 | |
| GSE47630 | ADAM17 | 6868 | 7 | 0 | 33 | |
| GSE54993 | ADAM17 | 6868 | 0 | 7 | 63 | |
| GSE54994 | ADAM17 | 6868 | 11 | 1 | 41 | |
| GSE60625 | ADAM17 | 6868 | 0 | 3 | 8 | |
| GSE74703 | ADAM17 | 6868 | 2 | 0 | 34 | |
| GSE74704 | ADAM17 | 6868 | 9 | 1 | 10 | |
| TCGA | ADAM17 | 6868 | 34 | 5 | 57 |
Total number of gains: 90; Total number of losses: 26; Total Number of normals: 372.
Somatic mutations of ADAM17:
Generating mutation plots.
Highly correlated genes for ADAM17:
Showing top 20/1500 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ADAM17 | PSME4 | 0.793699 | 11 | 0 | 11 |
| ADAM17 | EME1 | 0.769989 | 5 | 0 | 4 |
| ADAM17 | NFKB2 | 0.768312 | 3 | 0 | 3 |
| ADAM17 | FAM89A | 0.767717 | 3 | 0 | 3 |
| ADAM17 | RNF213 | 0.764619 | 3 | 0 | 3 |
| ADAM17 | COMMD2 | 0.761214 | 4 | 0 | 4 |
| ADAM17 | SPRR2G | 0.760276 | 3 | 0 | 3 |
| ADAM17 | PDE8A | 0.734146 | 3 | 0 | 3 |
| ADAM17 | BLACAT1 | 0.731719 | 4 | 0 | 4 |
| ADAM17 | MMS22L | 0.730904 | 3 | 0 | 3 |
| ADAM17 | CPSF3 | 0.728994 | 8 | 0 | 7 |
| ADAM17 | HAS3 | 0.728757 | 5 | 0 | 5 |
| ADAM17 | XPR1 | 0.726581 | 7 | 0 | 6 |
| ADAM17 | CEP55 | 0.725259 | 10 | 0 | 9 |
| ADAM17 | CAD | 0.721408 | 11 | 0 | 11 |
| ADAM17 | ISG20L2 | 0.71665 | 10 | 0 | 9 |
| ADAM17 | MPLKIP | 0.71655 | 6 | 0 | 6 |
| ADAM17 | LRRC8A | 0.712344 | 3 | 0 | 3 |
| ADAM17 | NEK8 | 0.712003 | 3 | 0 | 3 |
| ADAM17 | RFWD3 | 0.711297 | 10 | 0 | 10 |
For details and further investigation, click here