| Full name: ATPase H+ transporting V0 subunit c | Alias Symbol: VATL|Vma3 | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 527 | HGNC ID: HGNC:855 | Ensembl Gene: ENSG00000185883 | OMIM ID: 108745 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V0C involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05152 | Tuberculosis |
Expression of ATP6V0C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V0C | 527 | 36994_at | -0.1353 | 0.7223 | |
| GSE20347 | ATP6V0C | 527 | 36994_at | -0.5279 | 0.0000 | |
| GSE23400 | ATP6V0C | 527 | 36994_at | -0.3393 | 0.0000 | |
| GSE26886 | ATP6V0C | 527 | 36994_at | -0.6219 | 0.0052 | |
| GSE29001 | ATP6V0C | 527 | 36994_at | -0.2454 | 0.2309 | |
| GSE38129 | ATP6V0C | 527 | 36994_at | -0.2368 | 0.0509 | |
| GSE45670 | ATP6V0C | 527 | 36994_at | -0.0105 | 0.9473 | |
| GSE63941 | ATP6V0C | 527 | 36994_at | 0.2242 | 0.3989 | |
| GSE77861 | ATP6V0C | 527 | 36994_at | -0.2978 | 0.2422 | |
| GSE97050 | ATP6V0C | 527 | A_23_P54816 | -0.1171 | 0.8062 | |
| SRP007169 | ATP6V0C | 527 | RNAseq | -1.5206 | 0.0000 | |
| SRP008496 | ATP6V0C | 527 | RNAseq | -1.7787 | 0.0000 | |
| SRP064894 | ATP6V0C | 527 | RNAseq | -0.6263 | 0.0032 | |
| SRP133303 | ATP6V0C | 527 | RNAseq | -0.3941 | 0.0140 | |
| SRP159526 | ATP6V0C | 527 | RNAseq | -0.6544 | 0.0432 | |
| SRP193095 | ATP6V0C | 527 | RNAseq | -0.6652 | 0.0005 | |
| SRP219564 | ATP6V0C | 527 | RNAseq | -0.0774 | 0.8130 | |
| TCGA | ATP6V0C | 527 | RNAseq | 0.0500 | 0.2820 |
Upregulated datasets: 0; Downregulated datasets: 2.
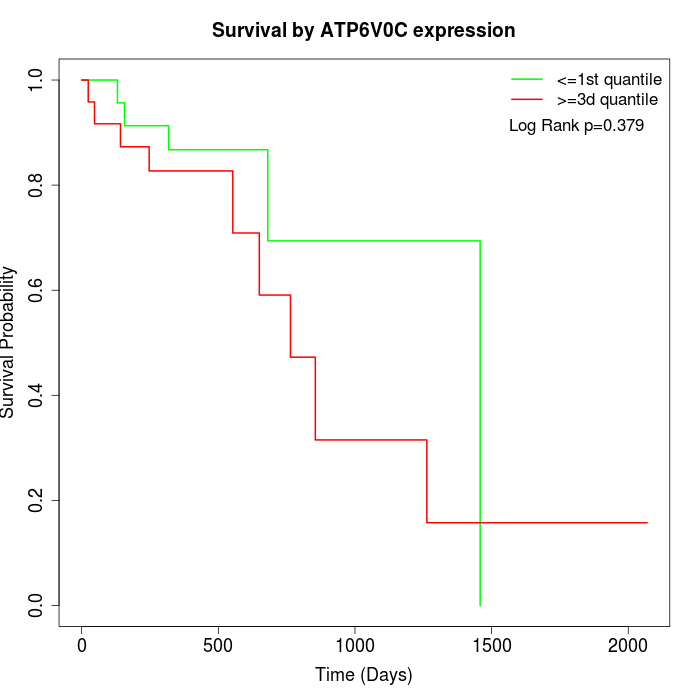
Survival by ATP6V0C expression:
Note: Click image to view full size file.
Copy number change of ATP6V0C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V0C | 527 | 5 | 5 | 20 | |
| GSE20123 | ATP6V0C | 527 | 5 | 4 | 21 | |
| GSE43470 | ATP6V0C | 527 | 4 | 7 | 32 | |
| GSE46452 | ATP6V0C | 527 | 38 | 1 | 20 | |
| GSE47630 | ATP6V0C | 527 | 13 | 6 | 21 | |
| GSE54993 | ATP6V0C | 527 | 3 | 5 | 62 | |
| GSE54994 | ATP6V0C | 527 | 5 | 9 | 39 | |
| GSE60625 | ATP6V0C | 527 | 4 | 0 | 7 | |
| GSE74703 | ATP6V0C | 527 | 4 | 5 | 27 | |
| GSE74704 | ATP6V0C | 527 | 3 | 2 | 15 | |
| TCGA | ATP6V0C | 527 | 20 | 13 | 63 |
Total number of gains: 104; Total number of losses: 57; Total Number of normals: 327.
Somatic mutations of ATP6V0C:
Generating mutation plots.
Highly correlated genes for ATP6V0C:
Showing top 20/907 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V0C | ZDHHC5 | 0.735778 | 3 | 0 | 3 |
| ATP6V0C | VPS25 | 0.731165 | 4 | 0 | 4 |
| ATP6V0C | DUOXA1 | 0.709474 | 4 | 0 | 4 |
| ATP6V0C | S100A16 | 0.706704 | 5 | 0 | 5 |
| ATP6V0C | MFSD5 | 0.705814 | 9 | 0 | 9 |
| ATP6V0C | CLVS1 | 0.698473 | 3 | 0 | 3 |
| ATP6V0C | MYO5B | 0.69282 | 4 | 0 | 3 |
| ATP6V0C | CSK | 0.680686 | 9 | 0 | 9 |
| ATP6V0C | RHOG | 0.68009 | 9 | 0 | 9 |
| ATP6V0C | RARG | 0.673212 | 9 | 0 | 7 |
| ATP6V0C | SPINT1 | 0.672637 | 9 | 0 | 8 |
| ATP6V0C | TOLLIP | 0.671409 | 8 | 0 | 6 |
| ATP6V0C | SH3GL1 | 0.668702 | 11 | 0 | 11 |
| ATP6V0C | GTF3C1 | 0.668532 | 10 | 0 | 9 |
| ATP6V0C | CARHSP1 | 0.662926 | 9 | 0 | 9 |
| ATP6V0C | EPHA1 | 0.662552 | 7 | 0 | 6 |
| ATP6V0C | SLC35C1 | 0.661959 | 8 | 0 | 8 |
| ATP6V0C | TOM1 | 0.656389 | 9 | 0 | 8 |
| ATP6V0C | RASSF5 | 0.655784 | 5 | 0 | 4 |
| ATP6V0C | PRSS8 | 0.655397 | 9 | 0 | 8 |
For details and further investigation, click here