| Full name: Fas associated via death domain | Alias Symbol: MORT1|GIG3 | ||
| Type: protein-coding gene | Cytoband: 11q13.3 | ||
| Entrez ID: 8772 | HGNC ID: HGNC:3573 | Ensembl Gene: ENSG00000168040 | OMIM ID: 602457 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
FADD involved pathways:
Expression of FADD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FADD | 8772 | 202535_at | 1.8387 | 0.1911 | |
| GSE20347 | FADD | 8772 | 202535_at | 1.7208 | 0.0003 | |
| GSE23400 | FADD | 8772 | 202535_at | 1.3072 | 0.0000 | |
| GSE26886 | FADD | 8772 | 202535_at | 1.6823 | 0.0001 | |
| GSE29001 | FADD | 8772 | 202535_at | 1.6489 | 0.0004 | |
| GSE38129 | FADD | 8772 | 202535_at | 1.7172 | 0.0000 | |
| GSE45670 | FADD | 8772 | 202535_at | 0.9709 | 0.0265 | |
| GSE63941 | FADD | 8772 | 202535_at | 1.8647 | 0.0467 | |
| GSE77861 | FADD | 8772 | 202535_at | 1.2146 | 0.1536 | |
| GSE97050 | FADD | 8772 | A_23_P86917 | 1.4471 | 0.0781 | |
| SRP007169 | FADD | 8772 | RNAseq | 1.7104 | 0.0167 | |
| SRP008496 | FADD | 8772 | RNAseq | 1.9485 | 0.0090 | |
| SRP064894 | FADD | 8772 | RNAseq | 2.2902 | 0.0000 | |
| SRP133303 | FADD | 8772 | RNAseq | 2.2655 | 0.0000 | |
| SRP159526 | FADD | 8772 | RNAseq | 2.9893 | 0.0000 | |
| SRP193095 | FADD | 8772 | RNAseq | 2.2798 | 0.0000 | |
| SRP219564 | FADD | 8772 | RNAseq | 2.6982 | 0.0001 | |
| TCGA | FADD | 8772 | RNAseq | 0.5806 | 0.0000 |
Upregulated datasets: 13; Downregulated datasets: 0.
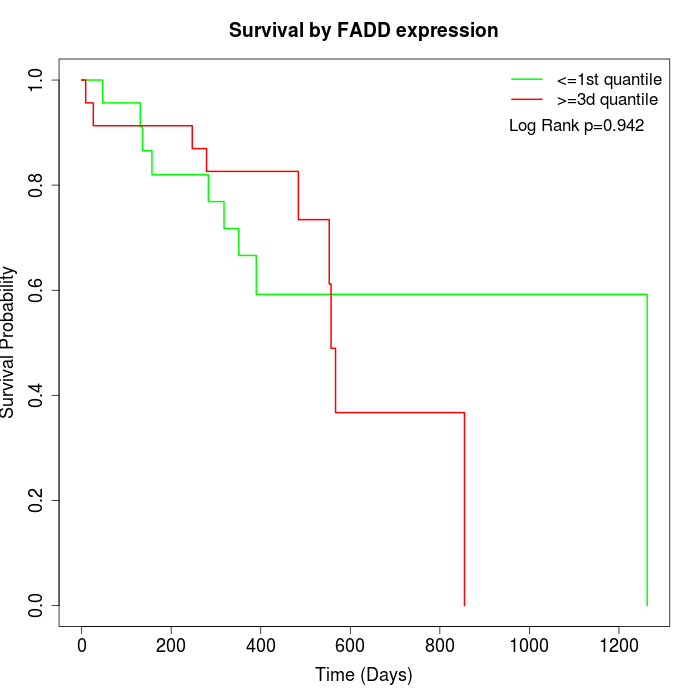
Survival by FADD expression:
Note: Click image to view full size file.
Copy number change of FADD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FADD | 8772 | 19 | 2 | 9 | |
| GSE20123 | FADD | 8772 | 20 | 1 | 9 | |
| GSE43470 | FADD | 8772 | 15 | 2 | 26 | |
| GSE46452 | FADD | 8772 | 33 | 3 | 23 | |
| GSE47630 | FADD | 8772 | 18 | 2 | 20 | |
| GSE54993 | FADD | 8772 | 1 | 25 | 44 | |
| GSE54994 | FADD | 8772 | 26 | 5 | 22 | |
| GSE60625 | FADD | 8772 | 0 | 8 | 3 | |
| GSE74703 | FADD | 8772 | 12 | 1 | 23 | |
| GSE74704 | FADD | 8772 | 13 | 0 | 7 | |
| TCGA | FADD | 8772 | 64 | 2 | 30 |
Total number of gains: 221; Total number of losses: 51; Total Number of normals: 216.
Somatic mutations of FADD:
Generating mutation plots.
Highly correlated genes for FADD:
Showing top 20/1458 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FADD | PPFIA1 | 0.90905 | 11 | 0 | 11 |
| FADD | ANO1 | 0.798028 | 11 | 0 | 11 |
| FADD | A4GALT | 0.755975 | 3 | 0 | 3 |
| FADD | ENPP5 | 0.755483 | 3 | 0 | 3 |
| FADD | MRPL21 | 0.755185 | 7 | 0 | 7 |
| FADD | BAD | 0.752885 | 3 | 0 | 3 |
| FADD | TRIM59 | 0.751581 | 3 | 0 | 3 |
| FADD | SPSB2 | 0.748287 | 3 | 0 | 3 |
| FADD | ASB7 | 0.747958 | 3 | 0 | 3 |
| FADD | FLVCR1 | 0.744481 | 3 | 0 | 3 |
| FADD | SPICE1 | 0.73209 | 3 | 0 | 3 |
| FADD | SAP130 | 0.729323 | 3 | 0 | 3 |
| FADD | ANLN | 0.729125 | 4 | 0 | 4 |
| FADD | DHRS13 | 0.724356 | 3 | 0 | 3 |
| FADD | ZDHHC16 | 0.724067 | 4 | 0 | 4 |
| FADD | CHML | 0.720176 | 3 | 0 | 3 |
| FADD | GTF2F2 | 0.715737 | 3 | 0 | 3 |
| FADD | MASTL | 0.711955 | 4 | 0 | 3 |
| FADD | UBTD2 | 0.711183 | 5 | 0 | 5 |
| FADD | CTTN | 0.709598 | 10 | 0 | 8 |
For details and further investigation, click here