| Full name: BAR/IMD domain containing adaptor protein 2 | Alias Symbol: BAP2|IRSp53 | ||
| Type: protein-coding gene | Cytoband: 17q25.3 | ||
| Entrez ID: 10458 | HGNC ID: HGNC:947 | Ensembl Gene: ENSG00000175866 | OMIM ID: 605475 |
| Drug and gene relationship at DGIdb | |||
BAIAP2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04520 | Adherens junction | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of BAIAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BAIAP2 | 10458 | 209502_s_at | -0.9180 | 0.1664 | |
| GSE20347 | BAIAP2 | 10458 | 209502_s_at | -0.9620 | 0.0000 | |
| GSE23400 | BAIAP2 | 10458 | 209502_s_at | -0.4712 | 0.0000 | |
| GSE26886 | BAIAP2 | 10458 | 209502_s_at | -0.9628 | 0.0001 | |
| GSE29001 | BAIAP2 | 10458 | 205294_at | -0.7083 | 0.0246 | |
| GSE38129 | BAIAP2 | 10458 | 209502_s_at | -0.5864 | 0.0263 | |
| GSE45670 | BAIAP2 | 10458 | 209502_s_at | -0.1588 | 0.4310 | |
| GSE53622 | BAIAP2 | 10458 | 18601 | -0.9354 | 0.0000 | |
| GSE53624 | BAIAP2 | 10458 | 18601 | -0.9402 | 0.0000 | |
| GSE63941 | BAIAP2 | 10458 | 205293_x_at | 1.3031 | 0.0014 | |
| GSE77861 | BAIAP2 | 10458 | 209502_s_at | -0.2737 | 0.4398 | |
| GSE97050 | BAIAP2 | 10458 | A_24_P159648 | -0.0984 | 0.8918 | |
| SRP007169 | BAIAP2 | 10458 | RNAseq | -2.0421 | 0.0000 | |
| SRP008496 | BAIAP2 | 10458 | RNAseq | -1.8138 | 0.0000 | |
| SRP064894 | BAIAP2 | 10458 | RNAseq | -1.4393 | 0.0000 | |
| SRP133303 | BAIAP2 | 10458 | RNAseq | -0.9230 | 0.0029 | |
| SRP159526 | BAIAP2 | 10458 | RNAseq | -1.0972 | 0.0102 | |
| SRP193095 | BAIAP2 | 10458 | RNAseq | -0.7223 | 0.0040 | |
| SRP219564 | BAIAP2 | 10458 | RNAseq | -0.9344 | 0.0555 | |
| TCGA | BAIAP2 | 10458 | RNAseq | 0.0549 | 0.5196 |
Upregulated datasets: 1; Downregulated datasets: 4.
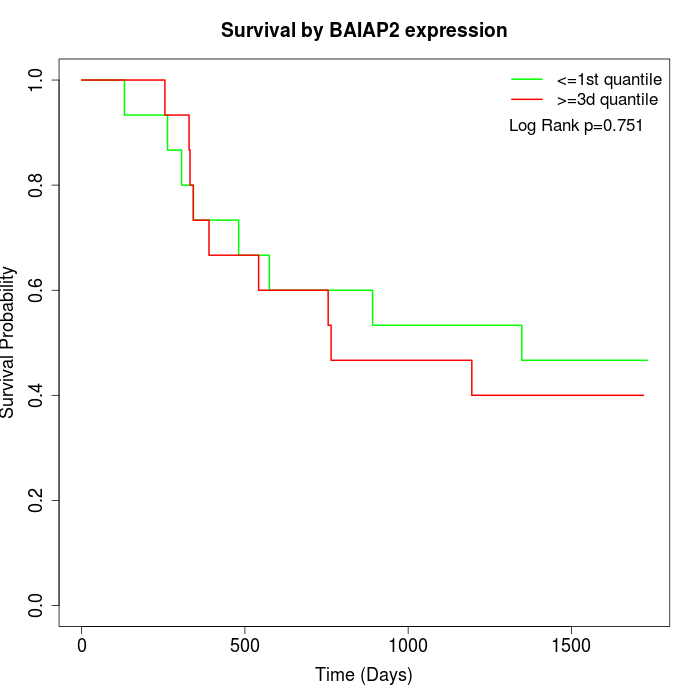
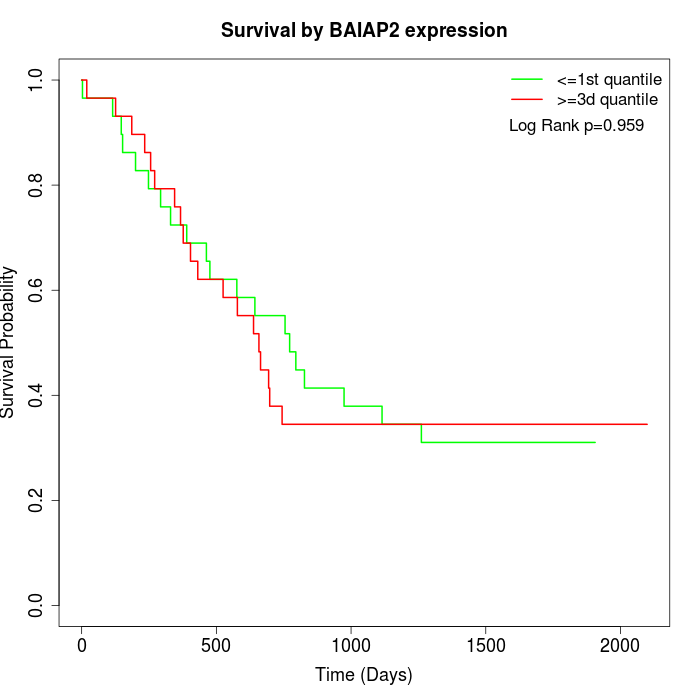
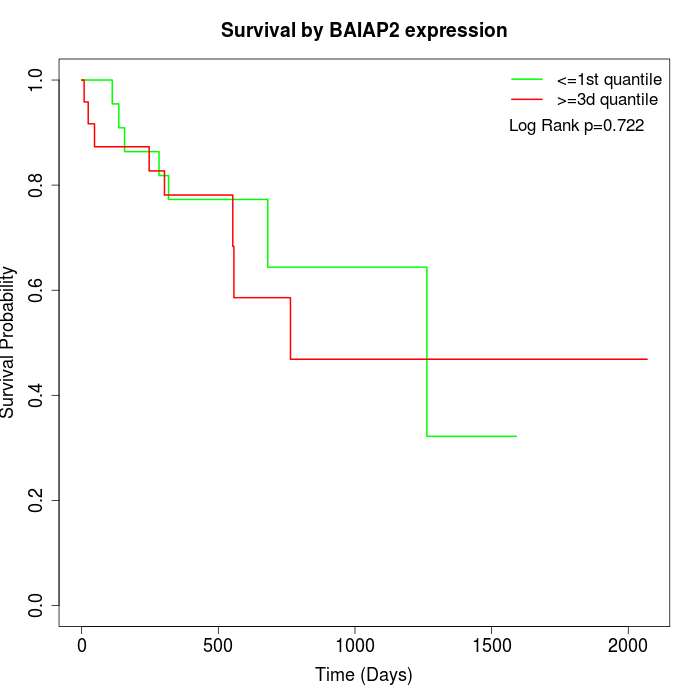
Survival by BAIAP2 expression:
Note: Click image to view full size file.
Copy number change of BAIAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BAIAP2 | 10458 | 4 | 4 | 22 | |
| GSE20123 | BAIAP2 | 10458 | 4 | 4 | 22 | |
| GSE43470 | BAIAP2 | 10458 | 3 | 5 | 35 | |
| GSE46452 | BAIAP2 | 10458 | 34 | 0 | 25 | |
| GSE47630 | BAIAP2 | 10458 | 9 | 2 | 29 | |
| GSE54993 | BAIAP2 | 10458 | 2 | 5 | 63 | |
| GSE54994 | BAIAP2 | 10458 | 10 | 6 | 37 | |
| GSE60625 | BAIAP2 | 10458 | 6 | 0 | 5 | |
| GSE74703 | BAIAP2 | 10458 | 3 | 3 | 30 | |
| GSE74704 | BAIAP2 | 10458 | 4 | 3 | 13 | |
| TCGA | BAIAP2 | 10458 | 27 | 13 | 56 |
Total number of gains: 106; Total number of losses: 45; Total Number of normals: 337.
Somatic mutations of BAIAP2:
Generating mutation plots.
Highly correlated genes for BAIAP2:
Showing top 20/986 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BAIAP2 | TOM1 | 0.751301 | 10 | 0 | 10 |
| BAIAP2 | SH3GL1 | 0.740366 | 10 | 0 | 10 |
| BAIAP2 | OTOP3 | 0.735947 | 3 | 0 | 3 |
| BAIAP2 | TMEM45B | 0.728414 | 5 | 0 | 5 |
| BAIAP2 | IL18 | 0.724757 | 10 | 0 | 10 |
| BAIAP2 | C15orf48 | 0.723724 | 5 | 0 | 5 |
| BAIAP2 | EVPL | 0.721563 | 13 | 0 | 12 |
| BAIAP2 | RNF222 | 0.718559 | 3 | 0 | 3 |
| BAIAP2 | MALL | 0.717641 | 12 | 0 | 11 |
| BAIAP2 | TYRO3 | 0.715207 | 11 | 0 | 11 |
| BAIAP2 | RNF39 | 0.70863 | 9 | 0 | 9 |
| BAIAP2 | FAM163B | 0.7069 | 3 | 0 | 3 |
| BAIAP2 | LIPH | 0.706265 | 6 | 0 | 6 |
| BAIAP2 | PIM1 | 0.703246 | 10 | 0 | 9 |
| BAIAP2 | SPINK7 | 0.702767 | 6 | 0 | 5 |
| BAIAP2 | MFSD2B | 0.702234 | 3 | 0 | 3 |
| BAIAP2 | RAET1E | 0.702041 | 6 | 0 | 5 |
| BAIAP2 | MUC22 | 0.701563 | 3 | 0 | 3 |
| BAIAP2 | SIRT7 | 0.698768 | 12 | 0 | 10 |
| BAIAP2 | TMPRSS11E | 0.696494 | 10 | 0 | 9 |
For details and further investigation, click here