| Full name: biliverdin reductase A | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7p13 | ||
| Entrez ID: 644 | HGNC ID: HGNC:1062 | Ensembl Gene: ENSG00000106605 | OMIM ID: 109750 |
| Drug and gene relationship at DGIdb | |||
Expression of BLVRA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BLVRA | 644 | 211729_x_at | -0.6461 | 0.2526 | |
| GSE20347 | BLVRA | 644 | 211729_x_at | -0.6443 | 0.0001 | |
| GSE23400 | BLVRA | 644 | 203773_x_at | -0.3576 | 0.0000 | |
| GSE26886 | BLVRA | 644 | 203773_x_at | 0.1608 | 0.4293 | |
| GSE29001 | BLVRA | 644 | 211729_x_at | -0.6652 | 0.0011 | |
| GSE38129 | BLVRA | 644 | 211729_x_at | -0.4135 | 0.0016 | |
| GSE45670 | BLVRA | 644 | 203773_x_at | -0.5480 | 0.0010 | |
| GSE53622 | BLVRA | 644 | 62783 | -0.6350 | 0.0000 | |
| GSE53624 | BLVRA | 644 | 62783 | -0.5701 | 0.0000 | |
| GSE63941 | BLVRA | 644 | 211729_x_at | -0.0798 | 0.8952 | |
| GSE77861 | BLVRA | 644 | 211729_x_at | -0.6405 | 0.0334 | |
| GSE97050 | BLVRA | 644 | A_23_P71148 | -0.7265 | 0.0764 | |
| SRP007169 | BLVRA | 644 | RNAseq | -2.1083 | 0.0000 | |
| SRP008496 | BLVRA | 644 | RNAseq | -1.3367 | 0.0009 | |
| SRP064894 | BLVRA | 644 | RNAseq | -0.7855 | 0.0049 | |
| SRP133303 | BLVRA | 644 | RNAseq | -0.6341 | 0.0011 | |
| SRP159526 | BLVRA | 644 | RNAseq | -0.3978 | 0.1479 | |
| SRP193095 | BLVRA | 644 | RNAseq | -0.8974 | 0.0001 | |
| SRP219564 | BLVRA | 644 | RNAseq | -0.7075 | 0.0073 | |
| TCGA | BLVRA | 644 | RNAseq | 0.2050 | 0.0014 |
Upregulated datasets: 0; Downregulated datasets: 2.
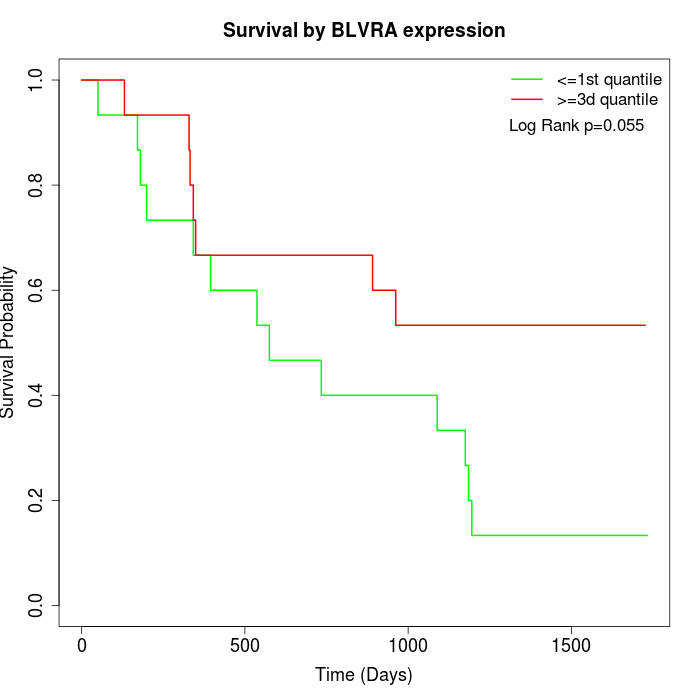
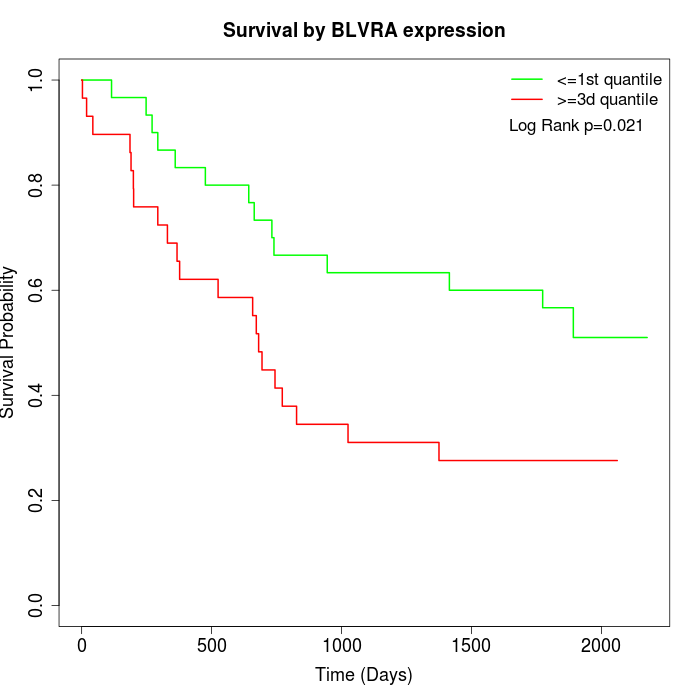
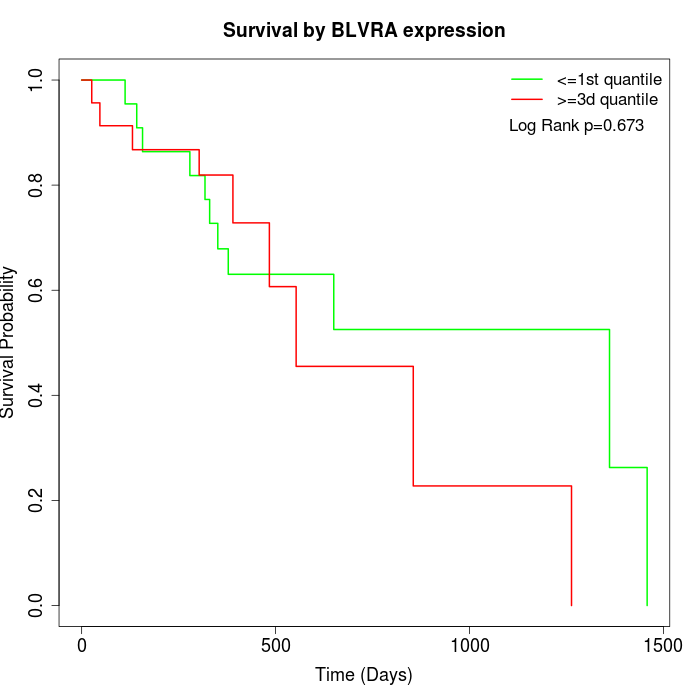
Survival by BLVRA expression:
Note: Click image to view full size file.
Copy number change of BLVRA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BLVRA | 644 | 14 | 0 | 16 | |
| GSE20123 | BLVRA | 644 | 13 | 0 | 17 | |
| GSE43470 | BLVRA | 644 | 4 | 0 | 39 | |
| GSE46452 | BLVRA | 644 | 11 | 1 | 47 | |
| GSE47630 | BLVRA | 644 | 8 | 1 | 31 | |
| GSE54993 | BLVRA | 644 | 0 | 8 | 62 | |
| GSE54994 | BLVRA | 644 | 17 | 3 | 33 | |
| GSE60625 | BLVRA | 644 | 0 | 0 | 11 | |
| GSE74703 | BLVRA | 644 | 4 | 0 | 32 | |
| GSE74704 | BLVRA | 644 | 7 | 0 | 13 | |
| TCGA | BLVRA | 644 | 50 | 7 | 39 |
Total number of gains: 128; Total number of losses: 20; Total Number of normals: 340.
Somatic mutations of BLVRA:
Generating mutation plots.
Highly correlated genes for BLVRA:
Showing top 20/1046 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BLVRA | LEPROT | 0.76352 | 3 | 0 | 3 |
| BLVRA | RSPH3 | 0.729509 | 4 | 0 | 4 |
| BLVRA | DBNL | 0.711137 | 3 | 0 | 3 |
| BLVRA | SAMD5 | 0.697399 | 6 | 0 | 6 |
| BLVRA | TAX1BP1 | 0.696044 | 12 | 0 | 11 |
| BLVRA | SLC46A2 | 0.692379 | 6 | 0 | 6 |
| BLVRA | EHD3 | 0.689888 | 10 | 0 | 10 |
| BLVRA | GNAT2 | 0.685111 | 3 | 0 | 3 |
| BLVRA | GYS2 | 0.681068 | 9 | 0 | 9 |
| BLVRA | ATP6V1D | 0.679609 | 9 | 0 | 7 |
| BLVRA | LMNA | 0.678945 | 4 | 0 | 4 |
| BLVRA | TMEM120A | 0.677426 | 5 | 0 | 5 |
| BLVRA | ZNF844 | 0.677192 | 4 | 0 | 4 |
| BLVRA | PMM1 | 0.677191 | 9 | 0 | 9 |
| BLVRA | ACOX3 | 0.675941 | 9 | 0 | 9 |
| BLVRA | CAST | 0.675679 | 10 | 0 | 9 |
| BLVRA | VAT1 | 0.673204 | 10 | 0 | 10 |
| BLVRA | SC5D | 0.672062 | 4 | 0 | 3 |
| BLVRA | GANC | 0.668855 | 6 | 0 | 5 |
| BLVRA | TRIP10 | 0.668816 | 10 | 0 | 10 |
For details and further investigation, click here