| Full name: calcium activated nucleotidase 1 | Alias Symbol: SHAPY|SCAN-1 | ||
| Type: protein-coding gene | Cytoband: 17q25.3 | ||
| Entrez ID: 124583 | HGNC ID: HGNC:19721 | Ensembl Gene: ENSG00000171302 | OMIM ID: 613165 |
| Drug and gene relationship at DGIdb | |||
Expression of CANT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CANT1 | 124583 | 221732_at | 0.1473 | 0.8584 | |
| GSE20347 | CANT1 | 124583 | 221732_at | -0.5124 | 0.0018 | |
| GSE23400 | CANT1 | 124583 | 221732_at | -0.2151 | 0.0049 | |
| GSE26886 | CANT1 | 124583 | 221732_at | -0.7730 | 0.0021 | |
| GSE29001 | CANT1 | 124583 | 221732_at | -0.5365 | 0.0022 | |
| GSE38129 | CANT1 | 124583 | 221732_at | -0.1790 | 0.2242 | |
| GSE45670 | CANT1 | 124583 | 46323_at | -0.1058 | 0.5607 | |
| GSE53622 | CANT1 | 124583 | 56124 | -0.3365 | 0.0003 | |
| GSE53624 | CANT1 | 124583 | 163312 | -0.5945 | 0.0000 | |
| GSE63941 | CANT1 | 124583 | 221732_at | 0.0000 | 0.9999 | |
| GSE77861 | CANT1 | 124583 | 221732_at | -0.3046 | 0.1778 | |
| GSE97050 | CANT1 | 124583 | A_23_P26759 | 0.1070 | 0.8465 | |
| SRP007169 | CANT1 | 124583 | RNAseq | -0.8189 | 0.0663 | |
| SRP008496 | CANT1 | 124583 | RNAseq | -0.9297 | 0.0002 | |
| SRP064894 | CANT1 | 124583 | RNAseq | -0.4040 | 0.0791 | |
| SRP133303 | CANT1 | 124583 | RNAseq | -0.2424 | 0.1867 | |
| SRP159526 | CANT1 | 124583 | RNAseq | -0.5139 | 0.0015 | |
| SRP193095 | CANT1 | 124583 | RNAseq | -0.3717 | 0.0896 | |
| SRP219564 | CANT1 | 124583 | RNAseq | -0.8363 | 0.1229 | |
| TCGA | CANT1 | 124583 | RNAseq | -0.1059 | 0.0260 |
Upregulated datasets: 0; Downregulated datasets: 0.
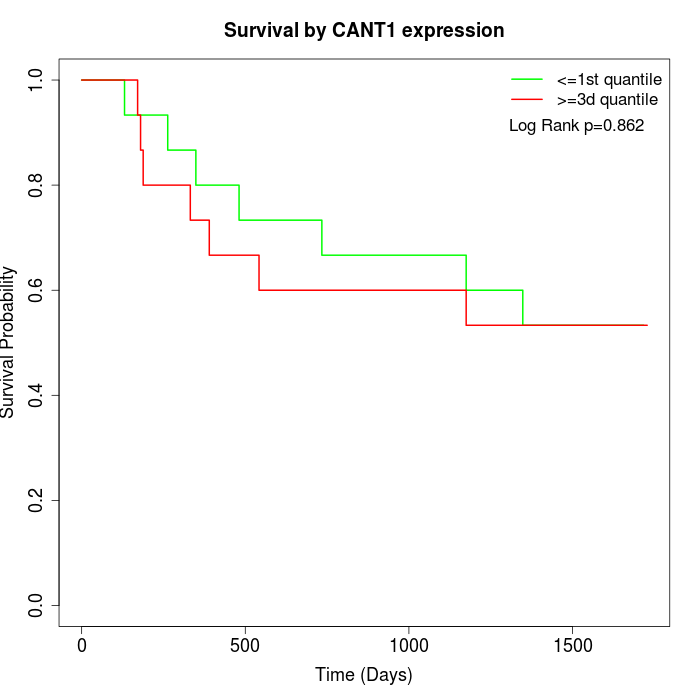
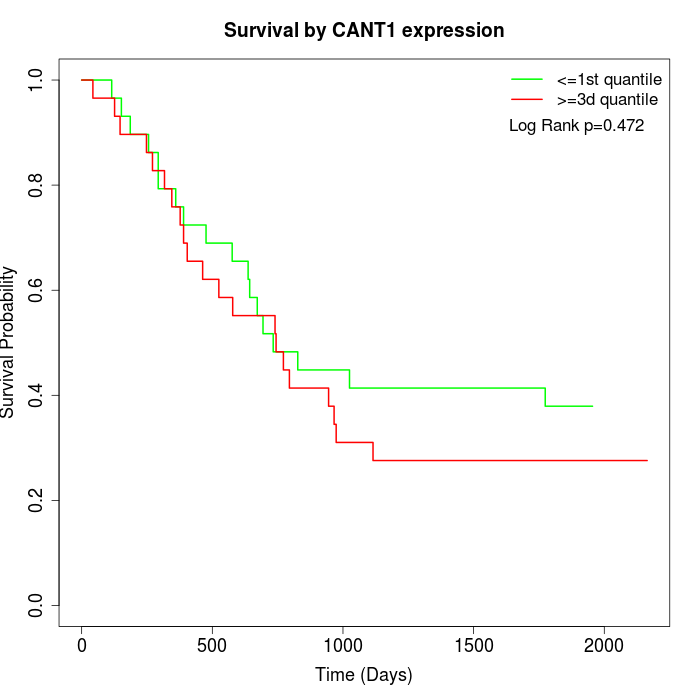
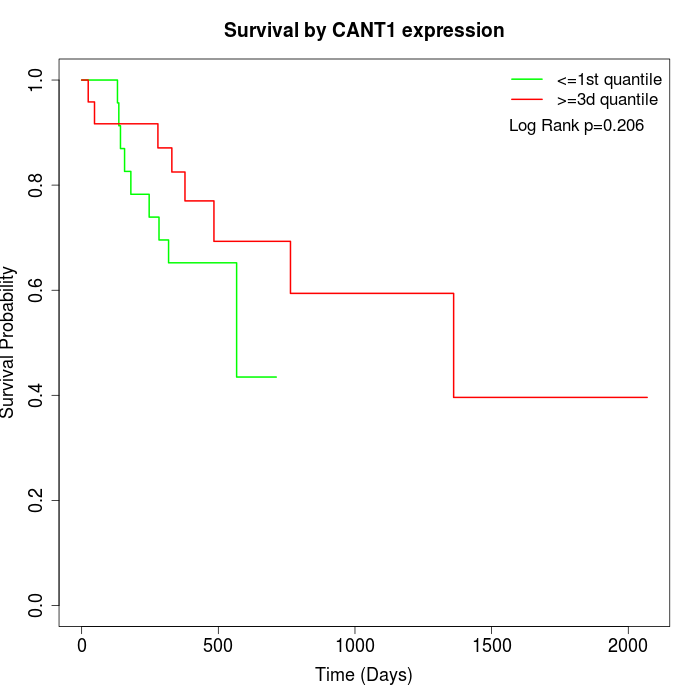
Survival by CANT1 expression:
Note: Click image to view full size file.
Copy number change of CANT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CANT1 | 124583 | 4 | 3 | 23 | |
| GSE20123 | CANT1 | 124583 | 4 | 3 | 23 | |
| GSE43470 | CANT1 | 124583 | 3 | 4 | 36 | |
| GSE46452 | CANT1 | 124583 | 33 | 0 | 26 | |
| GSE47630 | CANT1 | 124583 | 8 | 1 | 31 | |
| GSE54993 | CANT1 | 124583 | 2 | 5 | 63 | |
| GSE54994 | CANT1 | 124583 | 10 | 5 | 38 | |
| GSE60625 | CANT1 | 124583 | 6 | 0 | 5 | |
| GSE74703 | CANT1 | 124583 | 3 | 2 | 31 | |
| GSE74704 | CANT1 | 124583 | 4 | 3 | 13 | |
| TCGA | CANT1 | 124583 | 30 | 9 | 57 |
Total number of gains: 107; Total number of losses: 35; Total Number of normals: 346.
Somatic mutations of CANT1:
Generating mutation plots.
Highly correlated genes for CANT1:
Showing top 20/746 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CANT1 | DCAF12 | 0.765564 | 4 | 0 | 4 |
| CANT1 | CYP2C19 | 0.751549 | 3 | 0 | 3 |
| CANT1 | OSBPL8 | 0.728719 | 3 | 0 | 3 |
| CANT1 | CERS3 | 0.72023 | 3 | 0 | 3 |
| CANT1 | KIAA1522 | 0.715757 | 3 | 0 | 3 |
| CANT1 | KCNK6 | 0.715275 | 4 | 0 | 3 |
| CANT1 | PARG | 0.714369 | 3 | 0 | 3 |
| CANT1 | NIPAL1 | 0.705836 | 3 | 0 | 3 |
| CANT1 | NDST2 | 0.703945 | 3 | 0 | 3 |
| CANT1 | CIB1 | 0.699039 | 3 | 0 | 3 |
| CANT1 | DUSP18 | 0.686162 | 3 | 0 | 3 |
| CANT1 | SLCO2A1 | 0.680857 | 3 | 0 | 3 |
| CANT1 | AHRR | 0.675505 | 3 | 0 | 3 |
| CANT1 | E2F8 | 0.674556 | 4 | 0 | 3 |
| CANT1 | SLC39A11 | 0.674282 | 4 | 0 | 3 |
| CANT1 | DPM2 | 0.670924 | 3 | 0 | 3 |
| CANT1 | TET2 | 0.66955 | 4 | 0 | 3 |
| CANT1 | RNFT1 | 0.666582 | 3 | 0 | 3 |
| CANT1 | RNASEL | 0.665881 | 3 | 0 | 3 |
| CANT1 | TMEM86B | 0.665793 | 3 | 0 | 3 |
For details and further investigation, click here