| Full name: kinetochore localized astrin (SPAG5) binding protein | Alias Symbol: FLJ14502|SKAP|kinastrin|TRAF4AF1 | ||
| Type: protein-coding gene | Cytoband: 15q15.1 | ||
| Entrez ID: 90417 | HGNC ID: HGNC:30767 | Ensembl Gene: ENSG00000128944 | OMIM ID: 614718 |
| Drug and gene relationship at DGIdb | |||
Expression of KNSTRN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KNSTRN | 90417 | 225300_at | 0.8975 | 0.0559 | |
| GSE26886 | KNSTRN | 90417 | 225300_at | 1.8762 | 0.0000 | |
| GSE45670 | KNSTRN | 90417 | 225300_at | 1.1094 | 0.0002 | |
| GSE53622 | KNSTRN | 90417 | 3170 | 1.4638 | 0.0000 | |
| GSE53624 | KNSTRN | 90417 | 3170 | 1.9163 | 0.0000 | |
| GSE63941 | KNSTRN | 90417 | 225300_at | 1.7222 | 0.0021 | |
| GSE77861 | KNSTRN | 90417 | 225300_at | 0.8306 | 0.0690 | |
| SRP007169 | KNSTRN | 90417 | RNAseq | 2.0515 | 0.0000 | |
| SRP008496 | KNSTRN | 90417 | RNAseq | 1.6575 | 0.0000 | |
| SRP064894 | KNSTRN | 90417 | RNAseq | 1.3338 | 0.0000 | |
| SRP133303 | KNSTRN | 90417 | RNAseq | 1.6413 | 0.0000 | |
| SRP159526 | KNSTRN | 90417 | RNAseq | 1.6617 | 0.0000 | |
| SRP193095 | KNSTRN | 90417 | RNAseq | 1.1160 | 0.0000 | |
| SRP219564 | KNSTRN | 90417 | RNAseq | 0.8155 | 0.0416 |
Upregulated datasets: 11; Downregulated datasets: 0.
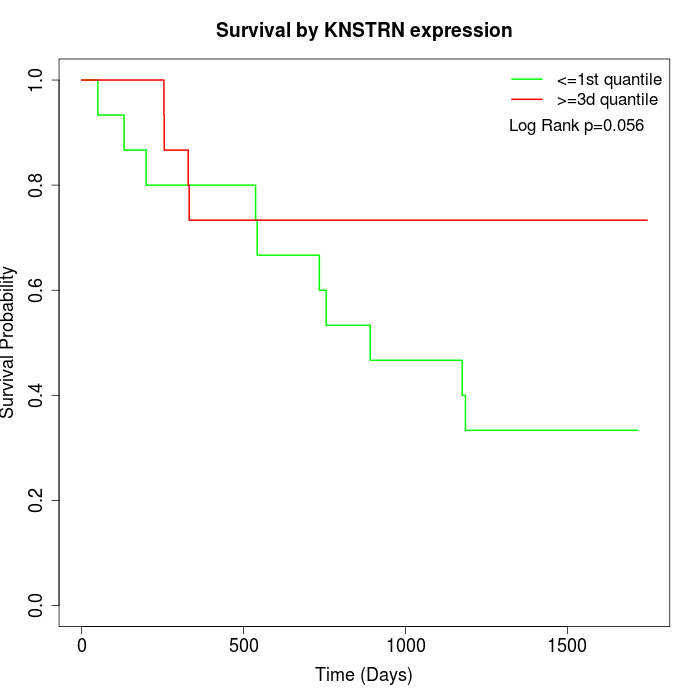
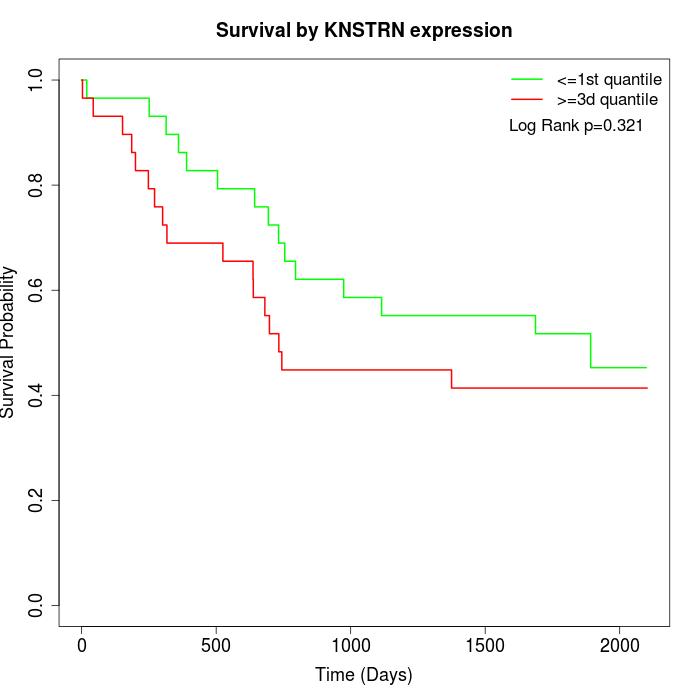
Survival by KNSTRN expression:
Note: Click image to view full size file.
Copy number change of KNSTRN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KNSTRN | 90417 | 4 | 5 | 21 | |
| GSE20123 | KNSTRN | 90417 | 4 | 5 | 21 | |
| GSE43470 | KNSTRN | 90417 | 4 | 6 | 33 | |
| GSE46452 | KNSTRN | 90417 | 3 | 7 | 49 | |
| GSE47630 | KNSTRN | 90417 | 8 | 11 | 21 | |
| GSE54993 | KNSTRN | 90417 | 4 | 6 | 60 | |
| GSE54994 | KNSTRN | 90417 | 5 | 7 | 41 | |
| GSE60625 | KNSTRN | 90417 | 4 | 0 | 7 | |
| GSE74703 | KNSTRN | 90417 | 4 | 5 | 27 | |
| GSE74704 | KNSTRN | 90417 | 3 | 3 | 14 | |
| TCGA | KNSTRN | 90417 | 11 | 17 | 68 |
Total number of gains: 54; Total number of losses: 72; Total Number of normals: 362.
Somatic mutations of KNSTRN:
Generating mutation plots.
Highly correlated genes for KNSTRN:
Showing top 20/1758 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KNSTRN | RAD51 | 0.87948 | 7 | 0 | 7 |
| KNSTRN | UBE2T | 0.863428 | 7 | 0 | 7 |
| KNSTRN | CCNB2 | 0.861009 | 6 | 0 | 6 |
| KNSTRN | BLM | 0.843468 | 7 | 0 | 7 |
| KNSTRN | NUSAP1 | 0.842302 | 7 | 0 | 7 |
| KNSTRN | PRC1 | 0.836238 | 7 | 0 | 7 |
| KNSTRN | ZWILCH | 0.8342 | 6 | 0 | 6 |
| KNSTRN | TRIP13 | 0.832307 | 7 | 0 | 7 |
| KNSTRN | NCAPH | 0.829799 | 5 | 0 | 5 |
| KNSTRN | CDCA5 | 0.829128 | 7 | 0 | 7 |
| KNSTRN | KIF23 | 0.829061 | 7 | 0 | 7 |
| KNSTRN | CDCA3 | 0.823196 | 7 | 0 | 7 |
| KNSTRN | UBE2C | 0.8231 | 7 | 0 | 7 |
| KNSTRN | KIF14 | 0.819832 | 7 | 0 | 7 |
| KNSTRN | FANCI | 0.816885 | 7 | 0 | 7 |
| KNSTRN | BUB1B | 0.816421 | 5 | 0 | 5 |
| KNSTRN | AURKB | 0.810509 | 7 | 0 | 7 |
| KNSTRN | SPAG5 | 0.810302 | 7 | 0 | 7 |
| KNSTRN | CDCA2 | 0.809383 | 5 | 0 | 5 |
| KNSTRN | TPX2 | 0.809207 | 7 | 0 | 7 |
For details and further investigation, click here