| Full name: C-C motif chemokine receptor 1 | Alias Symbol: CKR-1|MIP1aR|CD191 | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 1230 | HGNC ID: HGNC:1602 | Ensembl Gene: ENSG00000163823 | OMIM ID: 601159 |
| Drug and gene relationship at DGIdb | |||
CCR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCR1 | 1230 | 205099_s_at | 0.9345 | 0.3264 | |
| GSE20347 | CCR1 | 1230 | 205099_s_at | 0.8792 | 0.0004 | |
| GSE23400 | CCR1 | 1230 | 205098_at | 0.1578 | 0.0105 | |
| GSE26886 | CCR1 | 1230 | 205098_at | -0.2110 | 0.6023 | |
| GSE29001 | CCR1 | 1230 | 205098_at | 1.0375 | 0.0098 | |
| GSE38129 | CCR1 | 1230 | 205099_s_at | 0.7098 | 0.0004 | |
| GSE45670 | CCR1 | 1230 | 205098_at | 0.1426 | 0.7446 | |
| GSE53622 | CCR1 | 1230 | 114212 | 0.7181 | 0.0002 | |
| GSE53624 | CCR1 | 1230 | 114212 | 1.0487 | 0.0000 | |
| GSE63941 | CCR1 | 1230 | 205099_s_at | 0.0937 | 0.5939 | |
| GSE77861 | CCR1 | 1230 | 205099_s_at | 0.2065 | 0.0573 | |
| GSE97050 | CCR1 | 1230 | A_24_P148717 | 1.0166 | 0.1386 | |
| SRP007169 | CCR1 | 1230 | RNAseq | 3.6021 | 0.0000 | |
| SRP008496 | CCR1 | 1230 | RNAseq | 2.4899 | 0.0000 | |
| SRP064894 | CCR1 | 1230 | RNAseq | 0.9266 | 0.0226 | |
| SRP133303 | CCR1 | 1230 | RNAseq | 0.9954 | 0.0010 | |
| SRP159526 | CCR1 | 1230 | RNAseq | 0.8550 | 0.2771 | |
| SRP193095 | CCR1 | 1230 | RNAseq | 1.6334 | 0.0000 | |
| SRP219564 | CCR1 | 1230 | RNAseq | 0.8015 | 0.2950 | |
| TCGA | CCR1 | 1230 | RNAseq | 0.0482 | 0.7746 |
Upregulated datasets: 5; Downregulated datasets: 0.
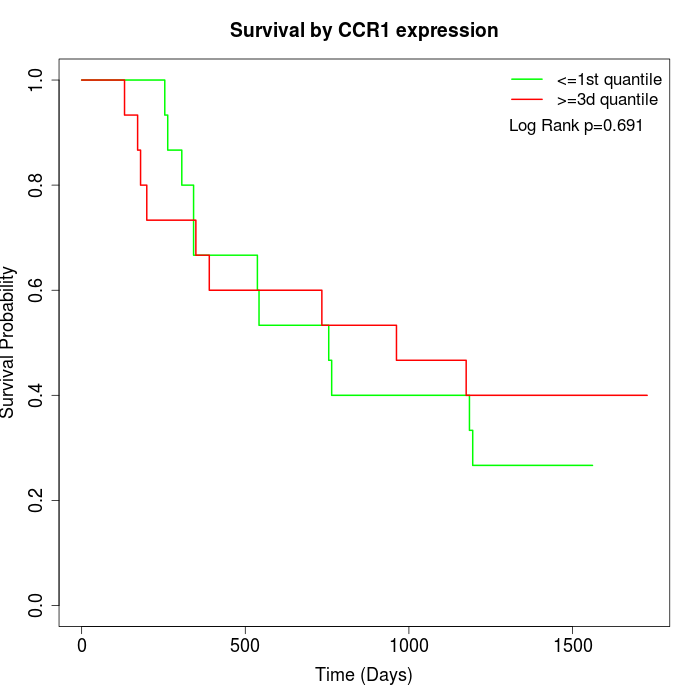
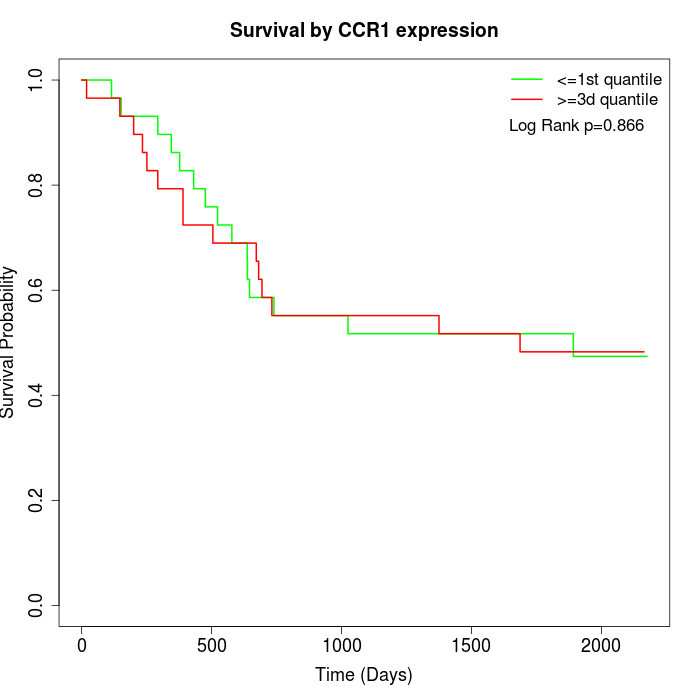
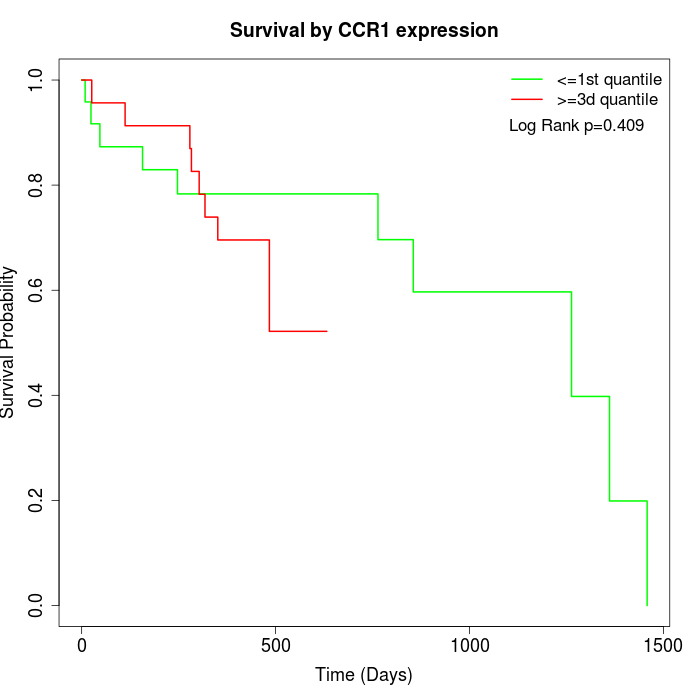
Survival by CCR1 expression:
Note: Click image to view full size file.
Copy number change of CCR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCR1 | 1230 | 0 | 17 | 13 | |
| GSE20123 | CCR1 | 1230 | 0 | 18 | 12 | |
| GSE43470 | CCR1 | 1230 | 0 | 20 | 23 | |
| GSE46452 | CCR1 | 1230 | 2 | 17 | 40 | |
| GSE47630 | CCR1 | 1230 | 2 | 23 | 15 | |
| GSE54993 | CCR1 | 1230 | 7 | 2 | 61 | |
| GSE54994 | CCR1 | 1230 | 0 | 36 | 17 | |
| GSE60625 | CCR1 | 1230 | 5 | 0 | 6 | |
| GSE74703 | CCR1 | 1230 | 0 | 15 | 21 | |
| GSE74704 | CCR1 | 1230 | 0 | 12 | 8 | |
| TCGA | CCR1 | 1230 | 1 | 75 | 20 |
Total number of gains: 17; Total number of losses: 235; Total Number of normals: 236.
Somatic mutations of CCR1:
Generating mutation plots.
Highly correlated genes for CCR1:
Showing top 20/685 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCR1 | FCGR3A | 0.809154 | 3 | 0 | 3 |
| CCR1 | TFEC | 0.779838 | 10 | 0 | 9 |
| CCR1 | TYROBP | 0.758463 | 11 | 0 | 11 |
| CCR1 | PIK3R5 | 0.745054 | 5 | 0 | 5 |
| CCR1 | CD163 | 0.744645 | 10 | 0 | 10 |
| CCR1 | FCER1G | 0.739219 | 11 | 0 | 10 |
| CCR1 | CYBB | 0.734202 | 11 | 0 | 10 |
| CCR1 | C1QB | 0.730976 | 11 | 0 | 11 |
| CCR1 | HAVCR2 | 0.730155 | 5 | 0 | 5 |
| CCR1 | SLC7A7 | 0.724992 | 12 | 0 | 10 |
| CCR1 | LILRB2 | 0.722531 | 11 | 0 | 10 |
| CCR1 | MS4A7 | 0.718696 | 4 | 0 | 4 |
| CCR1 | C19orf38 | 0.717905 | 3 | 0 | 3 |
| CCR1 | PLA2G7 | 0.717149 | 12 | 0 | 11 |
| CCR1 | GJD3 | 0.714143 | 4 | 0 | 4 |
| CCR1 | CCR5 | 0.711016 | 9 | 0 | 9 |
| CCR1 | IGSF6 | 0.709433 | 11 | 0 | 9 |
| CCR1 | CD53 | 0.702711 | 9 | 0 | 9 |
| CCR1 | MPEG1 | 0.702446 | 6 | 0 | 6 |
| CCR1 | ITGB2 | 0.70155 | 11 | 0 | 10 |
For details and further investigation, click here