| Full name: citron rho-interacting serine/threonine kinase | Alias Symbol: KIAA0949|STK21|CRIK|CITK | ||
| Type: protein-coding gene | Cytoband: 12q24.23 | ||
| Entrez ID: 11113 | HGNC ID: HGNC:1985 | Ensembl Gene: ENSG00000122966 | OMIM ID: 605629 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CIT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | CIT | 11113 | 103665 | 0.7361 | 0.0000 | |
| GSE53624 | CIT | 11113 | 103665 | 1.2505 | 0.0000 | |
| GSE97050 | CIT | 11113 | A_23_P420551 | 0.2509 | 0.4725 | |
| SRP007169 | CIT | 11113 | RNAseq | 2.5379 | 0.0000 | |
| SRP008496 | CIT | 11113 | RNAseq | 1.8836 | 0.0000 | |
| SRP064894 | CIT | 11113 | RNAseq | 1.0285 | 0.0003 | |
| SRP133303 | CIT | 11113 | RNAseq | 1.1263 | 0.0000 | |
| SRP159526 | CIT | 11113 | RNAseq | 2.2638 | 0.0013 | |
| SRP193095 | CIT | 11113 | RNAseq | 1.0980 | 0.0000 | |
| SRP219564 | CIT | 11113 | RNAseq | 0.6229 | 0.0333 | |
| TCGA | CIT | 11113 | RNAseq | 0.2299 | 0.0012 |
Upregulated datasets: 7; Downregulated datasets: 0.
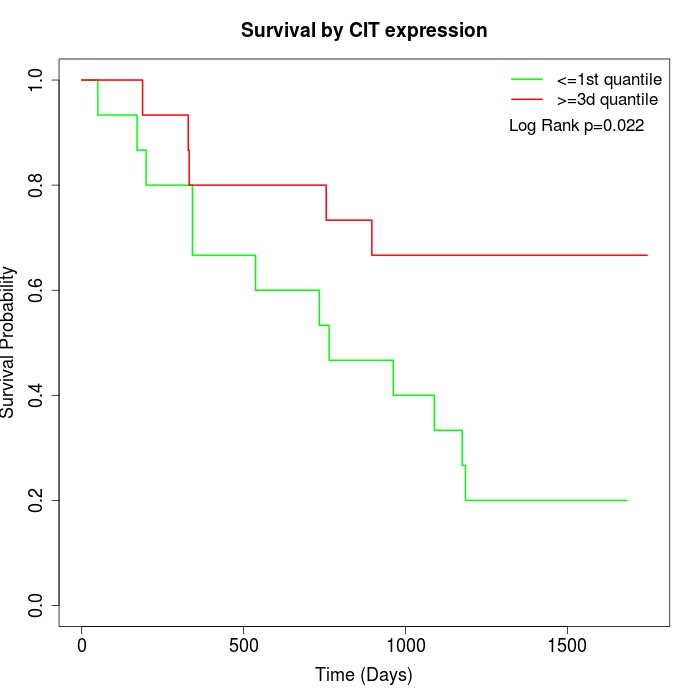
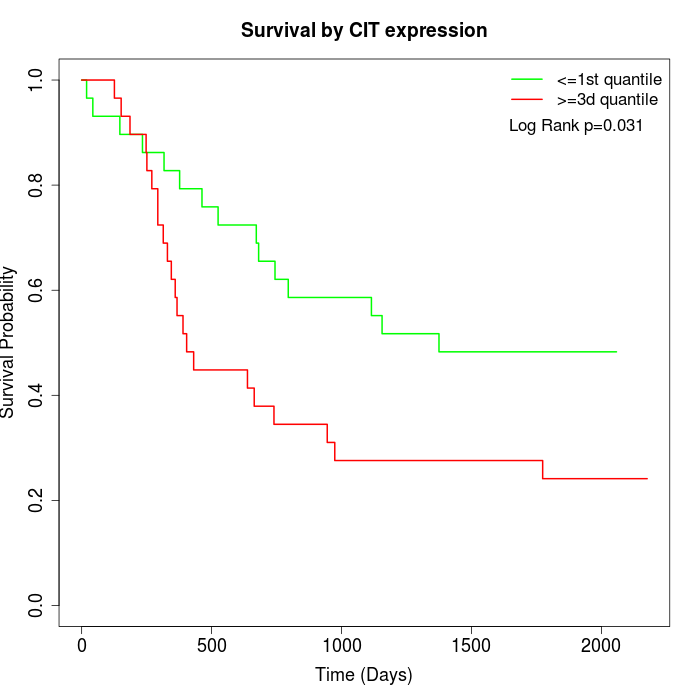
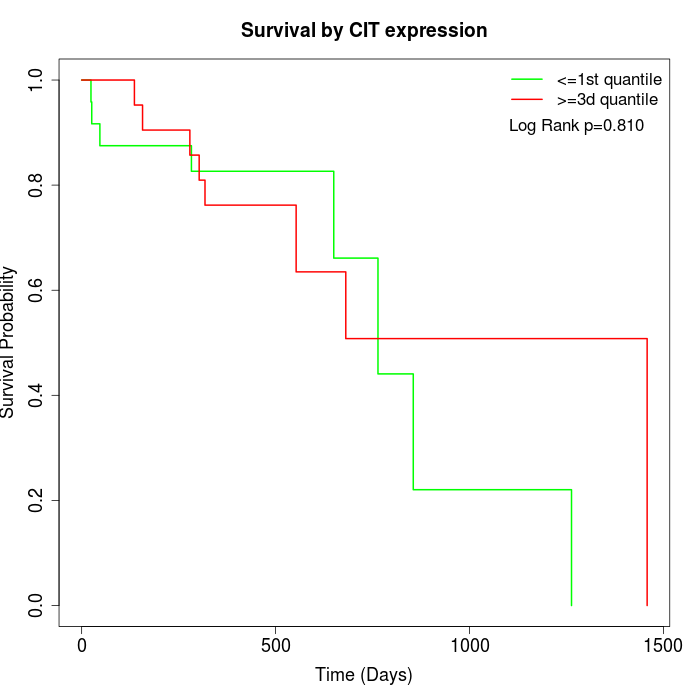
Survival by CIT expression:
Note: Click image to view full size file.
Copy number change of CIT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CIT | 11113 | 6 | 2 | 22 | |
| GSE20123 | CIT | 11113 | 6 | 2 | 22 | |
| GSE43470 | CIT | 11113 | 4 | 2 | 37 | |
| GSE46452 | CIT | 11113 | 9 | 1 | 49 | |
| GSE47630 | CIT | 11113 | 9 | 3 | 28 | |
| GSE54993 | CIT | 11113 | 0 | 5 | 65 | |
| GSE54994 | CIT | 11113 | 6 | 6 | 41 | |
| GSE60625 | CIT | 11113 | 0 | 0 | 11 | |
| GSE74703 | CIT | 11113 | 3 | 1 | 32 | |
| GSE74704 | CIT | 11113 | 3 | 2 | 15 | |
| TCGA | CIT | 11113 | 21 | 11 | 64 |
Total number of gains: 67; Total number of losses: 35; Total Number of normals: 386.
Somatic mutations of CIT:
Generating mutation plots.
Highly correlated genes for CIT:
Showing top 20/294 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CIT | BUB1 | 0.746333 | 3 | 0 | 3 |
| CIT | PIF1 | 0.728313 | 3 | 0 | 3 |
| CIT | GANAB | 0.724848 | 3 | 0 | 3 |
| CIT | NDC80 | 0.72035 | 3 | 0 | 3 |
| CIT | SNRPF | 0.716769 | 3 | 0 | 3 |
| CIT | MPHOSPH9 | 0.710953 | 3 | 0 | 3 |
| CIT | SMARCD1 | 0.70921 | 3 | 0 | 3 |
| CIT | CENPE | 0.70861 | 3 | 0 | 3 |
| CIT | ATAD2 | 0.707792 | 3 | 0 | 3 |
| CIT | PSMB4 | 0.704801 | 3 | 0 | 3 |
| CIT | NASP | 0.703605 | 3 | 0 | 3 |
| CIT | UBE2O | 0.702736 | 3 | 0 | 3 |
| CIT | BCL7A | 0.698726 | 3 | 0 | 3 |
| CIT | ANLN | 0.697965 | 3 | 0 | 3 |
| CIT | TIMELESS | 0.697509 | 3 | 0 | 3 |
| CIT | NCAPG2 | 0.696936 | 3 | 0 | 3 |
| CIT | GMPS | 0.693275 | 3 | 0 | 3 |
| CIT | PARL | 0.692826 | 3 | 0 | 3 |
| CIT | BRIP1 | 0.690994 | 3 | 0 | 3 |
| CIT | GINS1 | 0.690615 | 3 | 0 | 3 |
For details and further investigation, click here