| Full name: dehydrogenase/reductase 4 like 2 | Alias Symbol: SDR25C3 | ||
| Type: protein-coding gene | Cytoband: 14q11.2 | ||
| Entrez ID: 317749 | HGNC ID: HGNC:19731 | Ensembl Gene: ENSG00000187630 | OMIM ID: 615196 |
| Drug and gene relationship at DGIdb | |||
Expression of DHRS4L2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | DHRS4L2 | 317749 | 91597 | -0.8135 | 0.0000 | |
| GSE53624 | DHRS4L2 | 317749 | 91597 | -0.4678 | 0.0000 | |
| GSE97050 | DHRS4L2 | 317749 | A_33_P3213997 | -0.4779 | 0.1057 | |
| SRP007169 | DHRS4L2 | 317749 | RNAseq | -1.6230 | 0.0000 | |
| SRP008496 | DHRS4L2 | 317749 | RNAseq | -1.5937 | 0.0000 | |
| SRP064894 | DHRS4L2 | 317749 | RNAseq | 0.6354 | 0.1126 | |
| SRP133303 | DHRS4L2 | 317749 | RNAseq | -0.4611 | 0.1117 | |
| SRP159526 | DHRS4L2 | 317749 | RNAseq | 0.0788 | 0.7313 | |
| SRP193095 | DHRS4L2 | 317749 | RNAseq | -0.5058 | 0.0047 | |
| SRP219564 | DHRS4L2 | 317749 | RNAseq | -0.0314 | 0.9224 | |
| TCGA | DHRS4L2 | 317749 | RNAseq | 0.0251 | 0.8698 |
Upregulated datasets: 0; Downregulated datasets: 2.
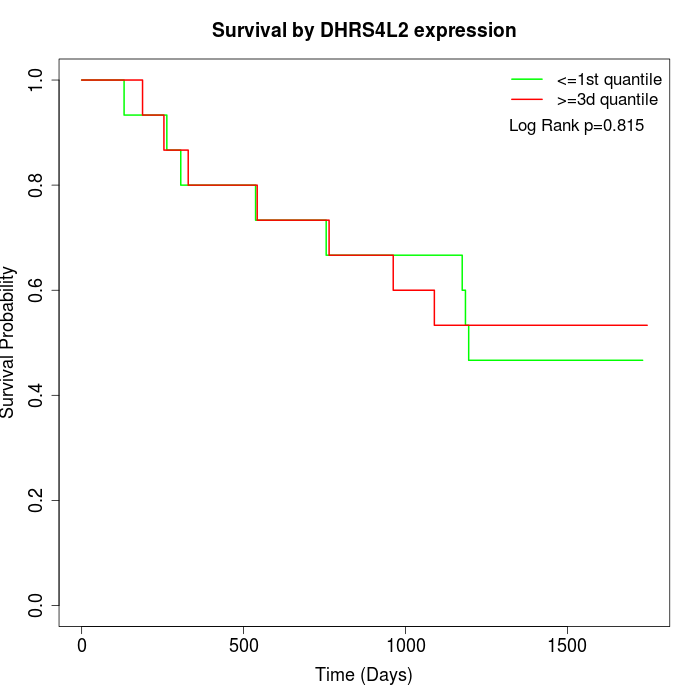
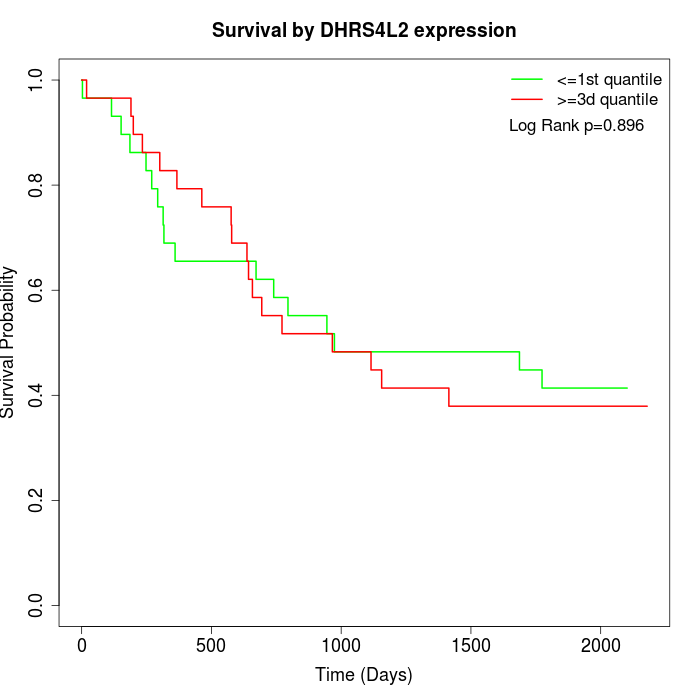
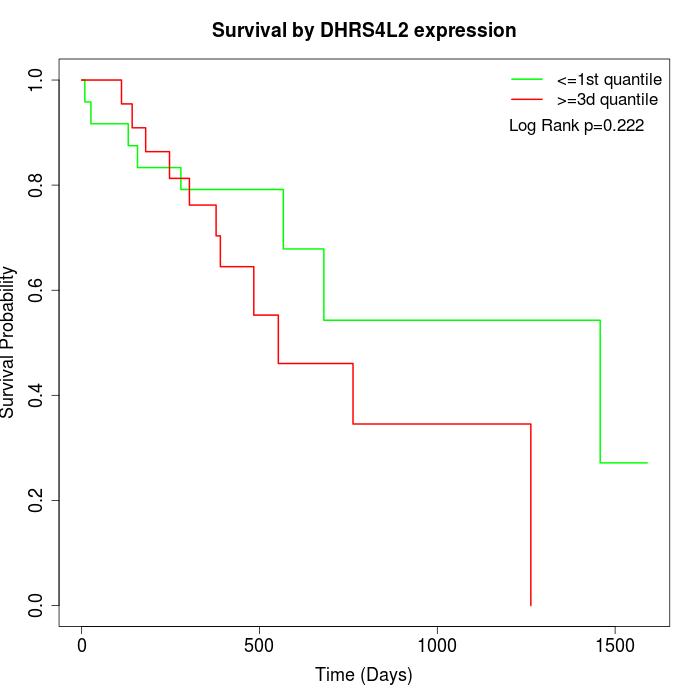
Survival by DHRS4L2 expression:
Note: Click image to view full size file.
Copy number change of DHRS4L2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DHRS4L2 | 317749 | 8 | 2 | 20 | |
| GSE20123 | DHRS4L2 | 317749 | 8 | 2 | 20 | |
| GSE43470 | DHRS4L2 | 317749 | 8 | 2 | 33 | |
| GSE46452 | DHRS4L2 | 317749 | 17 | 2 | 40 | |
| GSE47630 | DHRS4L2 | 317749 | 11 | 10 | 19 | |
| GSE54993 | DHRS4L2 | 317749 | 3 | 11 | 56 | |
| GSE54994 | DHRS4L2 | 317749 | 18 | 5 | 30 | |
| GSE60625 | DHRS4L2 | 317749 | 0 | 2 | 9 | |
| GSE74703 | DHRS4L2 | 317749 | 7 | 2 | 27 | |
| GSE74704 | DHRS4L2 | 317749 | 3 | 2 | 15 | |
| TCGA | DHRS4L2 | 317749 | 27 | 14 | 55 |
Total number of gains: 110; Total number of losses: 54; Total Number of normals: 324.
Somatic mutations of DHRS4L2:
Generating mutation plots.
Highly correlated genes for DHRS4L2:
Showing top 20/64 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DHRS4L2 | APEH | 0.733454 | 3 | 0 | 3 |
| DHRS4L2 | AP3B1 | 0.704685 | 3 | 0 | 3 |
| DHRS4L2 | FKBPL | 0.702236 | 3 | 0 | 3 |
| DHRS4L2 | PITHD1 | 0.700789 | 3 | 0 | 3 |
| DHRS4L2 | GUK1 | 0.70078 | 3 | 0 | 3 |
| DHRS4L2 | PRMT2 | 0.699976 | 3 | 0 | 3 |
| DHRS4L2 | MFSD5 | 0.693961 | 3 | 0 | 3 |
| DHRS4L2 | CCDC12 | 0.689693 | 3 | 0 | 3 |
| DHRS4L2 | GPR108 | 0.689422 | 3 | 0 | 3 |
| DHRS4L2 | SCAPER | 0.684831 | 3 | 0 | 3 |
| DHRS4L2 | EIF3K | 0.682061 | 3 | 0 | 3 |
| DHRS4L2 | ZBTB5 | 0.675259 | 3 | 0 | 3 |
| DHRS4L2 | FLII | 0.673094 | 3 | 0 | 3 |
| DHRS4L2 | COX6A1 | 0.670055 | 3 | 0 | 3 |
| DHRS4L2 | HK1 | 0.668616 | 3 | 0 | 3 |
| DHRS4L2 | FDX1 | 0.667465 | 3 | 0 | 3 |
| DHRS4L2 | PIGF | 0.662091 | 3 | 0 | 3 |
| DHRS4L2 | ALKBH7 | 0.661342 | 4 | 0 | 3 |
| DHRS4L2 | UBE2M | 0.65795 | 3 | 0 | 3 |
| DHRS4L2 | UNK | 0.655452 | 3 | 0 | 3 |
For details and further investigation, click here