| Full name: diaphanous related formin 3 | Alias Symbol: DRF3|FLJ34705|AN|NSDAN | ||
| Type: protein-coding gene | Cytoband: 13q21.2 | ||
| Entrez ID: 81624 | HGNC ID: HGNC:15480 | Ensembl Gene: ENSG00000139734 | OMIM ID: 614567 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
DIAPH3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04810 | Regulation of actin cytoskeleton |
Expression of DIAPH3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DIAPH3 | 81624 | 229097_at | 0.8807 | 0.2147 | |
| GSE20347 | DIAPH3 | 81624 | 220997_s_at | 0.0599 | 0.3556 | |
| GSE23400 | DIAPH3 | 81624 | 220997_s_at | 0.0271 | 0.5124 | |
| GSE26886 | DIAPH3 | 81624 | 229097_at | 0.3506 | 0.3087 | |
| GSE29001 | DIAPH3 | 81624 | 220997_s_at | 0.3959 | 0.0538 | |
| GSE38129 | DIAPH3 | 81624 | 220997_s_at | 0.1003 | 0.0567 | |
| GSE45670 | DIAPH3 | 81624 | 229097_at | 0.6765 | 0.0109 | |
| GSE53622 | DIAPH3 | 81624 | 17523 | 1.0885 | 0.0000 | |
| GSE53624 | DIAPH3 | 81624 | 120066 | 1.0507 | 0.0000 | |
| GSE63941 | DIAPH3 | 81624 | 229097_at | 0.7846 | 0.2987 | |
| GSE77861 | DIAPH3 | 81624 | 229097_at | 0.4316 | 0.2026 | |
| GSE97050 | DIAPH3 | 81624 | A_23_P162719 | 0.6551 | 0.2347 | |
| SRP007169 | DIAPH3 | 81624 | RNAseq | 1.4903 | 0.0019 | |
| SRP008496 | DIAPH3 | 81624 | RNAseq | 1.8006 | 0.0001 | |
| SRP064894 | DIAPH3 | 81624 | RNAseq | 0.8981 | 0.0055 | |
| SRP133303 | DIAPH3 | 81624 | RNAseq | 1.0859 | 0.0001 | |
| SRP159526 | DIAPH3 | 81624 | RNAseq | 0.4439 | 0.2685 | |
| SRP193095 | DIAPH3 | 81624 | RNAseq | 0.6752 | 0.0000 | |
| SRP219564 | DIAPH3 | 81624 | RNAseq | -0.0926 | 0.8481 | |
| TCGA | DIAPH3 | 81624 | RNAseq | 0.3063 | 0.0055 |
Upregulated datasets: 5; Downregulated datasets: 0.
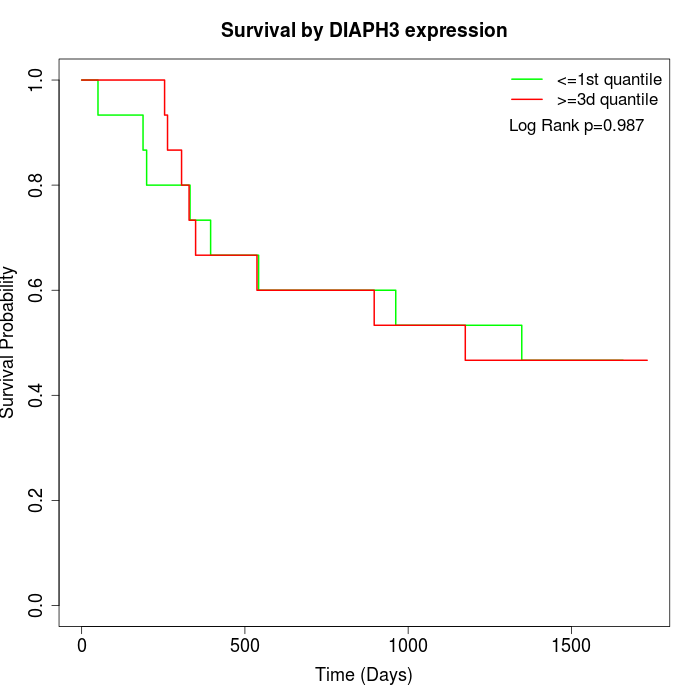
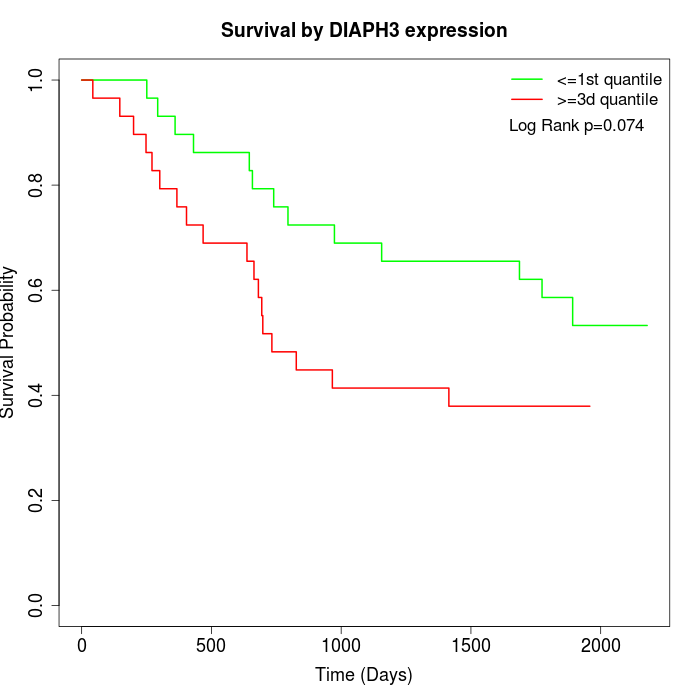
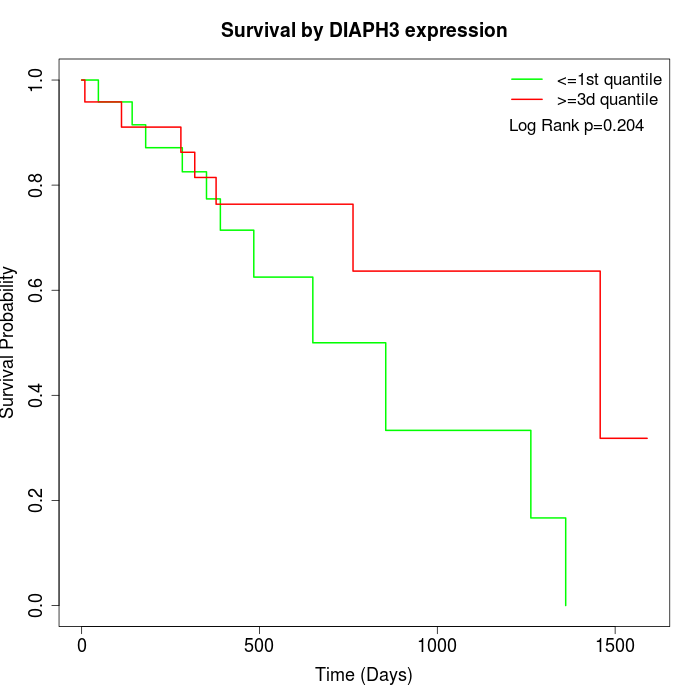
Survival by DIAPH3 expression:
Note: Click image to view full size file.
Copy number change of DIAPH3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DIAPH3 | 81624 | 2 | 11 | 17 | |
| GSE20123 | DIAPH3 | 81624 | 2 | 10 | 18 | |
| GSE43470 | DIAPH3 | 81624 | 1 | 12 | 30 | |
| GSE46452 | DIAPH3 | 81624 | 0 | 32 | 27 | |
| GSE47630 | DIAPH3 | 81624 | 2 | 26 | 12 | |
| GSE54993 | DIAPH3 | 81624 | 11 | 3 | 56 | |
| GSE54994 | DIAPH3 | 81624 | 3 | 13 | 37 | |
| GSE60625 | DIAPH3 | 81624 | 0 | 3 | 8 | |
| GSE74703 | DIAPH3 | 81624 | 1 | 10 | 25 | |
| GSE74704 | DIAPH3 | 81624 | 1 | 8 | 11 | |
| TCGA | DIAPH3 | 81624 | 11 | 35 | 50 |
Total number of gains: 34; Total number of losses: 163; Total Number of normals: 291.
Somatic mutations of DIAPH3:
Generating mutation plots.
Highly correlated genes for DIAPH3:
Showing top 20/589 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DIAPH3 | BOP1 | 0.754396 | 3 | 0 | 3 |
| DIAPH3 | DEPDC1B | 0.726101 | 8 | 0 | 8 |
| DIAPH3 | POC1A | 0.683653 | 8 | 0 | 8 |
| DIAPH3 | C19orf48 | 0.679576 | 3 | 0 | 3 |
| DIAPH3 | MARVELD3 | 0.673269 | 4 | 0 | 3 |
| DIAPH3 | LRWD1 | 0.67231 | 3 | 0 | 3 |
| DIAPH3 | NUF2 | 0.664863 | 8 | 0 | 7 |
| DIAPH3 | IRF6 | 0.664405 | 4 | 0 | 4 |
| DIAPH3 | ANLN | 0.660376 | 8 | 0 | 7 |
| DIAPH3 | CKAP2 | 0.660156 | 8 | 0 | 7 |
| DIAPH3 | RTKN | 0.659574 | 5 | 0 | 5 |
| DIAPH3 | TYMS | 0.656044 | 7 | 0 | 6 |
| DIAPH3 | CDCA5 | 0.654001 | 8 | 0 | 7 |
| DIAPH3 | POLD2 | 0.652268 | 4 | 0 | 3 |
| DIAPH3 | PYCR1 | 0.651355 | 4 | 0 | 4 |
| DIAPH3 | GART | 0.651296 | 4 | 0 | 4 |
| DIAPH3 | GPRIN1 | 0.649573 | 4 | 0 | 3 |
| DIAPH3 | FOLR3 | 0.649266 | 4 | 0 | 3 |
| DIAPH3 | HOXC10 | 0.648638 | 4 | 0 | 3 |
| DIAPH3 | SPC24 | 0.647863 | 7 | 0 | 6 |
For details and further investigation, click here