| Full name: ectodysplasin A receptor | Alias Symbol: ED5|EDA3|Edar|ED1R|EDA1R | ||
| Type: protein-coding gene | Cytoband: 2q13 | ||
| Entrez ID: 10913 | HGNC ID: HGNC:2895 | Ensembl Gene: ENSG00000135960 | OMIM ID: 604095 |
| Drug and gene relationship at DGIdb | |||
Expression of EDAR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EDAR | 10913 | 220048_at | -1.1893 | 0.0213 | |
| GSE20347 | EDAR | 10913 | 220048_at | -0.2272 | 0.2447 | |
| GSE23400 | EDAR | 10913 | 220048_at | -0.1064 | 0.0138 | |
| GSE26886 | EDAR | 10913 | 220048_at | -0.8305 | 0.0301 | |
| GSE29001 | EDAR | 10913 | 220048_at | -0.0332 | 0.9416 | |
| GSE38129 | EDAR | 10913 | 220048_at | 0.0167 | 0.9629 | |
| GSE45670 | EDAR | 10913 | 220048_at | -0.3389 | 0.0928 | |
| GSE53622 | EDAR | 10913 | 17941 | -0.7878 | 0.0273 | |
| GSE53624 | EDAR | 10913 | 17941 | -1.8873 | 0.0000 | |
| GSE63941 | EDAR | 10913 | 220048_at | 0.2253 | 0.6517 | |
| GSE77861 | EDAR | 10913 | 220048_at | -0.1806 | 0.3452 | |
| SRP007169 | EDAR | 10913 | RNAseq | -0.6202 | 0.5555 | |
| SRP064894 | EDAR | 10913 | RNAseq | -1.9030 | 0.0013 | |
| SRP133303 | EDAR | 10913 | RNAseq | -1.5197 | 0.0005 | |
| SRP159526 | EDAR | 10913 | RNAseq | -0.1734 | 0.8077 | |
| SRP193095 | EDAR | 10913 | RNAseq | -1.5393 | 0.0000 | |
| TCGA | EDAR | 10913 | RNAseq | 0.1232 | 0.7731 |
Upregulated datasets: 0; Downregulated datasets: 5.
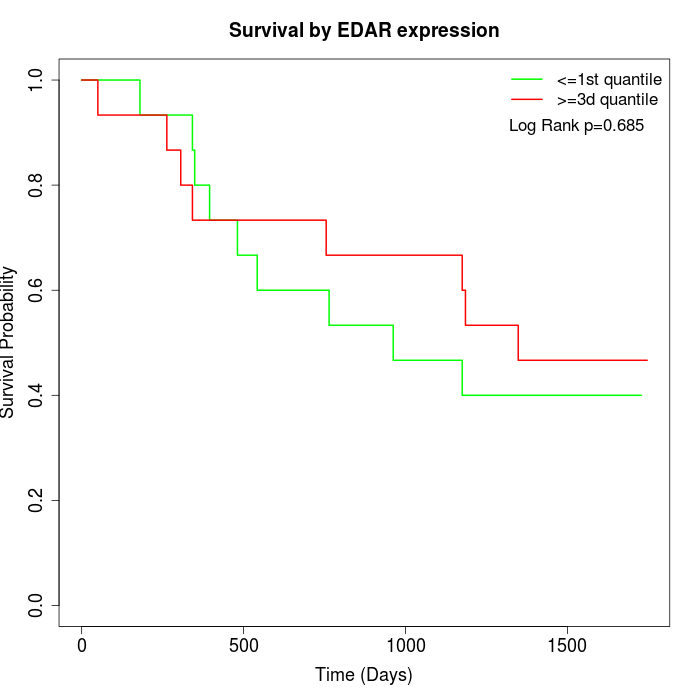
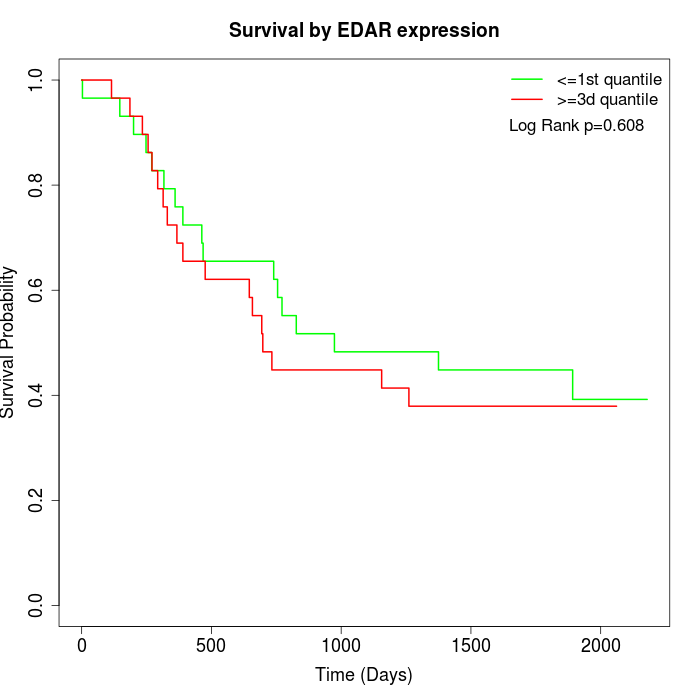
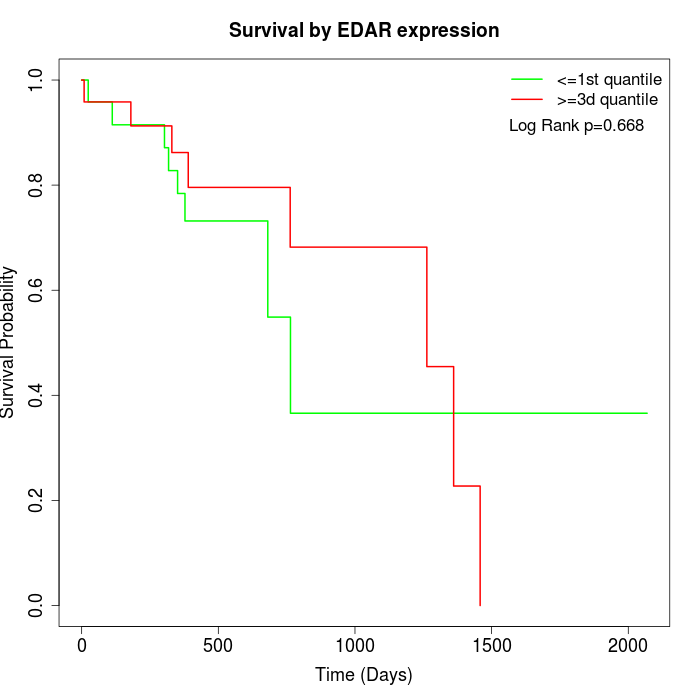
Survival by EDAR expression:
Note: Click image to view full size file.
Copy number change of EDAR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EDAR | 10913 | 4 | 2 | 24 | |
| GSE20123 | EDAR | 10913 | 4 | 2 | 24 | |
| GSE43470 | EDAR | 10913 | 3 | 1 | 39 | |
| GSE46452 | EDAR | 10913 | 1 | 4 | 54 | |
| GSE47630 | EDAR | 10913 | 7 | 0 | 33 | |
| GSE54993 | EDAR | 10913 | 0 | 6 | 64 | |
| GSE54994 | EDAR | 10913 | 10 | 0 | 43 | |
| GSE60625 | EDAR | 10913 | 0 | 3 | 8 | |
| GSE74703 | EDAR | 10913 | 3 | 1 | 32 | |
| GSE74704 | EDAR | 10913 | 2 | 2 | 16 | |
| TCGA | EDAR | 10913 | 31 | 6 | 59 |
Total number of gains: 65; Total number of losses: 27; Total Number of normals: 396.
Somatic mutations of EDAR:
Generating mutation plots.
Highly correlated genes for EDAR:
Showing top 20/217 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EDAR | CAPNS2 | 0.717896 | 3 | 0 | 3 |
| EDAR | GAPVD1 | 0.687697 | 3 | 0 | 3 |
| EDAR | AHNAK | 0.68626 | 3 | 0 | 3 |
| EDAR | CPT2 | 0.669045 | 3 | 0 | 3 |
| EDAR | FAR1 | 0.662555 | 3 | 0 | 3 |
| EDAR | PPP1CB | 0.633513 | 3 | 0 | 3 |
| EDAR | ENDOD1 | 0.62333 | 3 | 0 | 3 |
| EDAR | PPFIBP2 | 0.62153 | 6 | 0 | 4 |
| EDAR | RNF43 | 0.618026 | 4 | 0 | 3 |
| EDAR | ENDOU | 0.617444 | 4 | 0 | 3 |
| EDAR | SPAG17 | 0.615522 | 4 | 0 | 4 |
| EDAR | PDHB | 0.613723 | 3 | 0 | 3 |
| EDAR | PRKCE | 0.612846 | 4 | 0 | 3 |
| EDAR | MPP7 | 0.611411 | 5 | 0 | 3 |
| EDAR | RNF141 | 0.611359 | 5 | 0 | 4 |
| EDAR | PCYOX1 | 0.608464 | 5 | 0 | 5 |
| EDAR | SOX6 | 0.607265 | 5 | 0 | 3 |
| EDAR | PIK3CB | 0.60579 | 4 | 0 | 3 |
| EDAR | ARL6IP5 | 0.604217 | 4 | 0 | 3 |
| EDAR | SH3BGRL2 | 0.601303 | 5 | 0 | 4 |
For details and further investigation, click here