| Full name: enolase 1 | Alias Symbol: PPH|MBP-1 | ||
| Type: protein-coding gene | Cytoband: 1p36.23 | ||
| Entrez ID: 2023 | HGNC ID: HGNC:3350 | Ensembl Gene: ENSG00000074800 | OMIM ID: 172430 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ENO1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway |
Expression of ENO1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ENO1 | 2023 | 201231_s_at | 0.4618 | 0.2056 | |
| GSE20347 | ENO1 | 2023 | 201231_s_at | 0.4033 | 0.0138 | |
| GSE23400 | ENO1 | 2023 | 201231_s_at | 0.5933 | 0.0000 | |
| GSE26886 | ENO1 | 2023 | 201231_s_at | -0.4079 | 0.1241 | |
| GSE29001 | ENO1 | 2023 | 201231_s_at | 0.1297 | 0.8342 | |
| GSE38129 | ENO1 | 2023 | 201231_s_at | 0.8120 | 0.0000 | |
| GSE45670 | ENO1 | 2023 | 201231_s_at | 0.5145 | 0.0006 | |
| GSE53622 | ENO1 | 2023 | 1907 | 0.6391 | 0.0000 | |
| GSE53624 | ENO1 | 2023 | 1907 | 0.6955 | 0.0000 | |
| GSE63941 | ENO1 | 2023 | 201231_s_at | 1.4021 | 0.0008 | |
| GSE77861 | ENO1 | 2023 | 201231_s_at | 0.2354 | 0.5347 | |
| GSE97050 | ENO1 | 2023 | A_23_P137391 | 0.2729 | 0.3382 | |
| SRP007169 | ENO1 | 2023 | RNAseq | -0.3404 | 0.4732 | |
| SRP008496 | ENO1 | 2023 | RNAseq | -0.4084 | 0.1502 | |
| SRP064894 | ENO1 | 2023 | RNAseq | 0.7210 | 0.0004 | |
| SRP133303 | ENO1 | 2023 | RNAseq | 0.7965 | 0.0001 | |
| SRP159526 | ENO1 | 2023 | RNAseq | 0.3383 | 0.3711 | |
| SRP193095 | ENO1 | 2023 | RNAseq | 0.2468 | 0.2006 | |
| SRP219564 | ENO1 | 2023 | RNAseq | 0.2129 | 0.4378 | |
| TCGA | ENO1 | 2023 | RNAseq | 0.2924 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
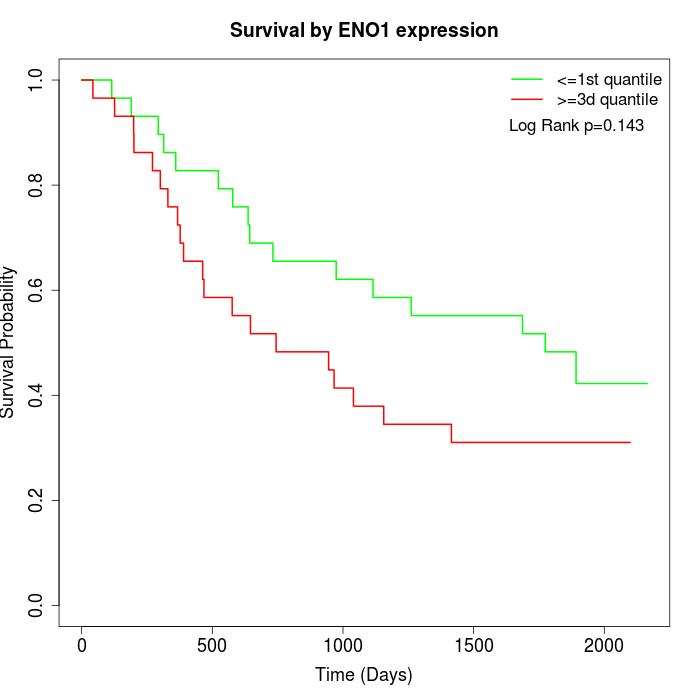
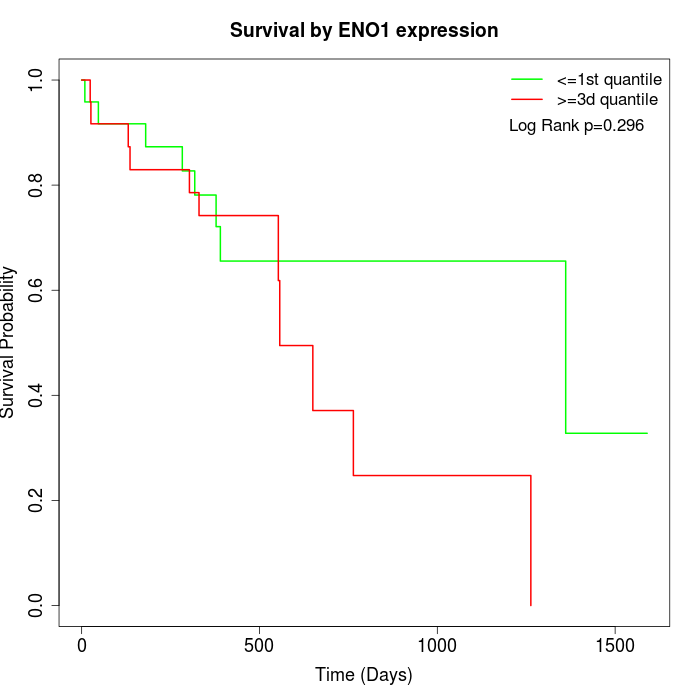
Survival by ENO1 expression:
Note: Click image to view full size file.
Copy number change of ENO1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ENO1 | 2023 | 1 | 5 | 24 | |
| GSE20123 | ENO1 | 2023 | 1 | 3 | 26 | |
| GSE43470 | ENO1 | 2023 | 6 | 5 | 32 | |
| GSE46452 | ENO1 | 2023 | 7 | 1 | 51 | |
| GSE47630 | ENO1 | 2023 | 8 | 4 | 28 | |
| GSE54993 | ENO1 | 2023 | 3 | 2 | 65 | |
| GSE54994 | ENO1 | 2023 | 12 | 4 | 37 | |
| GSE60625 | ENO1 | 2023 | 0 | 0 | 11 | |
| GSE74703 | ENO1 | 2023 | 5 | 3 | 28 | |
| GSE74704 | ENO1 | 2023 | 1 | 0 | 19 | |
| TCGA | ENO1 | 2023 | 11 | 24 | 61 |
Total number of gains: 55; Total number of losses: 51; Total Number of normals: 382.
Somatic mutations of ENO1:
Generating mutation plots.
Highly correlated genes for ENO1:
Showing top 20/816 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ENO1 | TSEN54 | 0.734856 | 3 | 0 | 3 |
| ENO1 | KIF2C | 0.729981 | 10 | 0 | 10 |
| ENO1 | CDC20 | 0.722861 | 10 | 0 | 10 |
| ENO1 | IDH3B | 0.719073 | 3 | 0 | 3 |
| ENO1 | LAMB3 | 0.707516 | 11 | 0 | 11 |
| ENO1 | MCAT | 0.704914 | 4 | 0 | 3 |
| ENO1 | DDOST | 0.697895 | 10 | 0 | 10 |
| ENO1 | PLK1 | 0.697006 | 10 | 0 | 9 |
| ENO1 | EBNA1BP2 | 0.686518 | 3 | 0 | 3 |
| ENO1 | STIL | 0.685322 | 10 | 0 | 9 |
| ENO1 | EFTUD2 | 0.679123 | 5 | 0 | 5 |
| ENO1 | MKI67 | 0.671108 | 10 | 0 | 9 |
| ENO1 | MRPS12 | 0.668727 | 6 | 0 | 6 |
| ENO1 | EMC1 | 0.668469 | 8 | 0 | 8 |
| ENO1 | RNASEH2A | 0.667956 | 11 | 0 | 10 |
| ENO1 | MAD2L1 | 0.665472 | 9 | 0 | 8 |
| ENO1 | CDKN3 | 0.665065 | 11 | 0 | 10 |
| ENO1 | KDM1A | 0.66002 | 10 | 0 | 10 |
| ENO1 | CCNA2 | 0.659545 | 9 | 0 | 9 |
| ENO1 | PIGU | 0.657201 | 5 | 0 | 4 |
For details and further investigation, click here