| Full name: polo like kinase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16p12.2 | ||
| Entrez ID: 5347 | HGNC ID: HGNC:9077 | Ensembl Gene: ENSG00000166851 | OMIM ID: 602098 |
| Drug and gene relationship at DGIdb | |||
PLK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04914 | Progesterone-mediated oocyte maturation |
Expression of PLK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLK1 | 5347 | 202240_at | 1.2339 | 0.1318 | |
| GSE20347 | PLK1 | 5347 | 202240_at | 0.9147 | 0.0000 | |
| GSE23400 | PLK1 | 5347 | 202240_at | 0.4893 | 0.0000 | |
| GSE26886 | PLK1 | 5347 | 202240_at | -0.0267 | 0.8807 | |
| GSE29001 | PLK1 | 5347 | 202240_at | 0.3240 | 0.0680 | |
| GSE38129 | PLK1 | 5347 | 202240_at | 0.9667 | 0.0000 | |
| GSE45670 | PLK1 | 5347 | 202240_at | 0.4842 | 0.0003 | |
| GSE53622 | PLK1 | 5347 | 61824 | 1.9819 | 0.0000 | |
| GSE53624 | PLK1 | 5347 | 15735 | 1.5418 | 0.0000 | |
| GSE63941 | PLK1 | 5347 | 202240_at | 3.7627 | 0.0000 | |
| GSE77861 | PLK1 | 5347 | 202240_at | 0.5218 | 0.0153 | |
| GSE97050 | PLK1 | 5347 | A_33_P3298387 | 1.9468 | 0.1133 | |
| SRP007169 | PLK1 | 5347 | RNAseq | 2.6527 | 0.0000 | |
| SRP008496 | PLK1 | 5347 | RNAseq | 2.2460 | 0.0000 | |
| SRP064894 | PLK1 | 5347 | RNAseq | 1.7842 | 0.0000 | |
| SRP133303 | PLK1 | 5347 | RNAseq | 1.8075 | 0.0000 | |
| SRP159526 | PLK1 | 5347 | RNAseq | 1.5711 | 0.0001 | |
| SRP193095 | PLK1 | 5347 | RNAseq | 1.2145 | 0.0000 | |
| SRP219564 | PLK1 | 5347 | RNAseq | 1.1975 | 0.0721 | |
| TCGA | PLK1 | 5347 | RNAseq | 1.0383 | 0.0000 |
Upregulated datasets: 10; Downregulated datasets: 0.
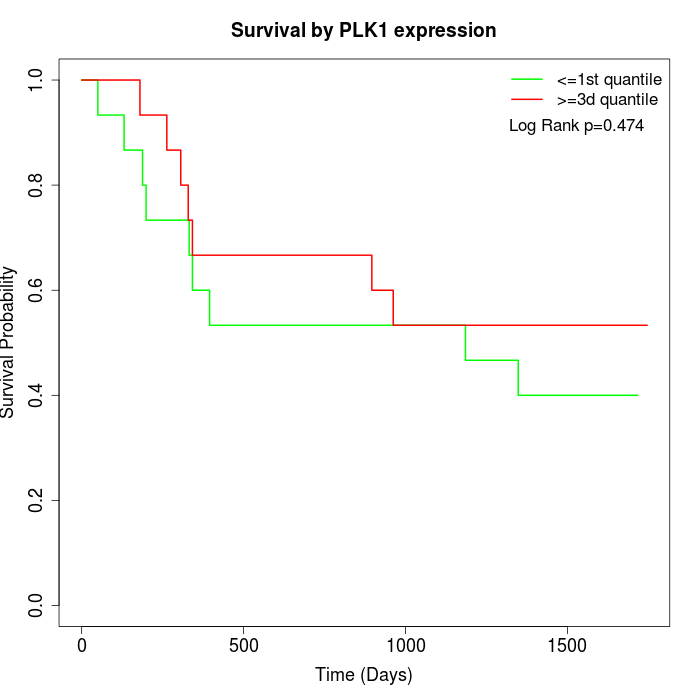
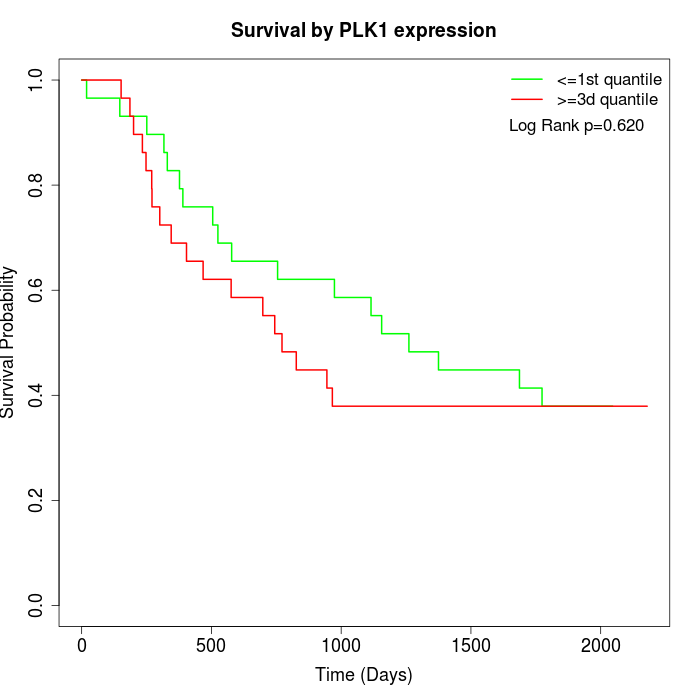
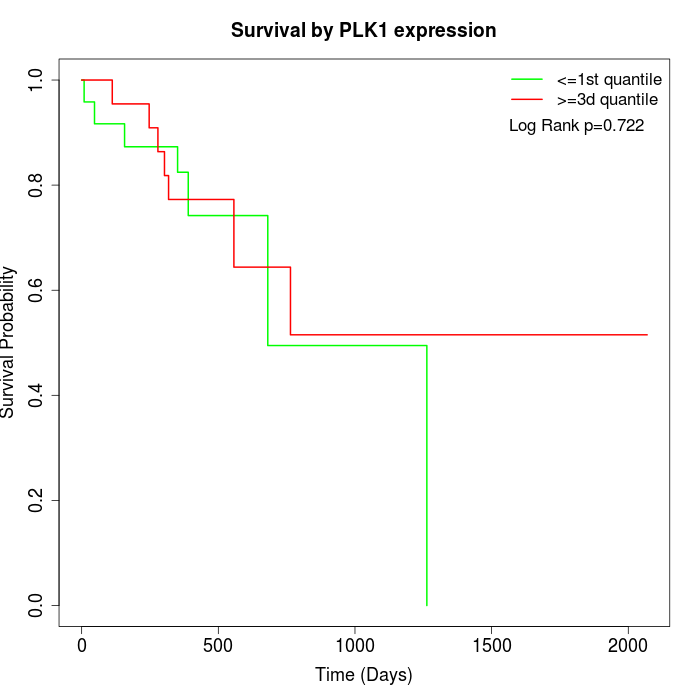
Survival by PLK1 expression:
Note: Click image to view full size file.
Copy number change of PLK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLK1 | 5347 | 6 | 4 | 20 | |
| GSE20123 | PLK1 | 5347 | 6 | 3 | 21 | |
| GSE43470 | PLK1 | 5347 | 3 | 3 | 37 | |
| GSE46452 | PLK1 | 5347 | 38 | 1 | 20 | |
| GSE47630 | PLK1 | 5347 | 11 | 6 | 23 | |
| GSE54993 | PLK1 | 5347 | 3 | 5 | 62 | |
| GSE54994 | PLK1 | 5347 | 4 | 8 | 41 | |
| GSE60625 | PLK1 | 5347 | 4 | 0 | 7 | |
| GSE74703 | PLK1 | 5347 | 3 | 2 | 31 | |
| GSE74704 | PLK1 | 5347 | 3 | 1 | 16 | |
| TCGA | PLK1 | 5347 | 19 | 13 | 64 |
Total number of gains: 100; Total number of losses: 46; Total Number of normals: 342.
Somatic mutations of PLK1:
Generating mutation plots.
Highly correlated genes for PLK1:
Showing top 20/1496 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLK1 | TPX2 | 0.83209 | 11 | 0 | 10 |
| PLK1 | UBE2T | 0.814212 | 7 | 0 | 7 |
| PLK1 | STIL | 0.812492 | 10 | 0 | 10 |
| PLK1 | CKS1B | 0.803714 | 11 | 0 | 11 |
| PLK1 | FOXM1 | 0.794533 | 11 | 0 | 11 |
| PLK1 | RNASEH2A | 0.793925 | 11 | 0 | 11 |
| PLK1 | KNSTRN | 0.790081 | 6 | 0 | 6 |
| PLK1 | PLK4 | 0.786248 | 8 | 0 | 8 |
| PLK1 | AURKB | 0.780725 | 12 | 0 | 11 |
| PLK1 | CENPW | 0.777591 | 7 | 0 | 7 |
| PLK1 | BIRC5 | 0.771867 | 11 | 0 | 10 |
| PLK1 | KIF14 | 0.770902 | 11 | 0 | 10 |
| PLK1 | MYBL2 | 0.770826 | 12 | 0 | 11 |
| PLK1 | CKAP2L | 0.770518 | 7 | 0 | 7 |
| PLK1 | CCNB2 | 0.769145 | 10 | 0 | 10 |
| PLK1 | KIF4A | 0.768275 | 10 | 0 | 9 |
| PLK1 | KPNA2 | 0.767959 | 8 | 0 | 8 |
| PLK1 | NUF2 | 0.767074 | 7 | 0 | 7 |
| PLK1 | EXO1 | 0.75836 | 11 | 0 | 10 |
| PLK1 | MKI67 | 0.758048 | 12 | 0 | 11 |
For details and further investigation, click here