| Full name: epsin 2 | Alias Symbol: KIAA1065|EHB21 | ||
| Type: protein-coding gene | Cytoband: 17p11.2 | ||
| Entrez ID: 22905 | HGNC ID: HGNC:18639 | Ensembl Gene: ENSG00000072134 | OMIM ID: 607263 |
| Drug and gene relationship at DGIdb | |||
Expression of EPN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | EPN2 | 22905 | 47490 | -0.4725 | 0.0000 | |
| GSE53624 | EPN2 | 22905 | 47490 | -0.4061 | 0.0000 | |
| GSE97050 | EPN2 | 22905 | A_23_P89310 | -0.3540 | 0.2974 | |
| SRP007169 | EPN2 | 22905 | RNAseq | -0.6624 | 0.2172 | |
| SRP008496 | EPN2 | 22905 | RNAseq | -0.3639 | 0.2135 | |
| SRP064894 | EPN2 | 22905 | RNAseq | -0.5671 | 0.0228 | |
| SRP133303 | EPN2 | 22905 | RNAseq | -0.7779 | 0.0000 | |
| SRP159526 | EPN2 | 22905 | RNAseq | -0.6832 | 0.0015 | |
| SRP193095 | EPN2 | 22905 | RNAseq | -0.6454 | 0.0006 | |
| SRP219564 | EPN2 | 22905 | RNAseq | -0.8339 | 0.0508 | |
| TCGA | EPN2 | 22905 | RNAseq | 0.1086 | 0.0476 |
Upregulated datasets: 0; Downregulated datasets: 0.
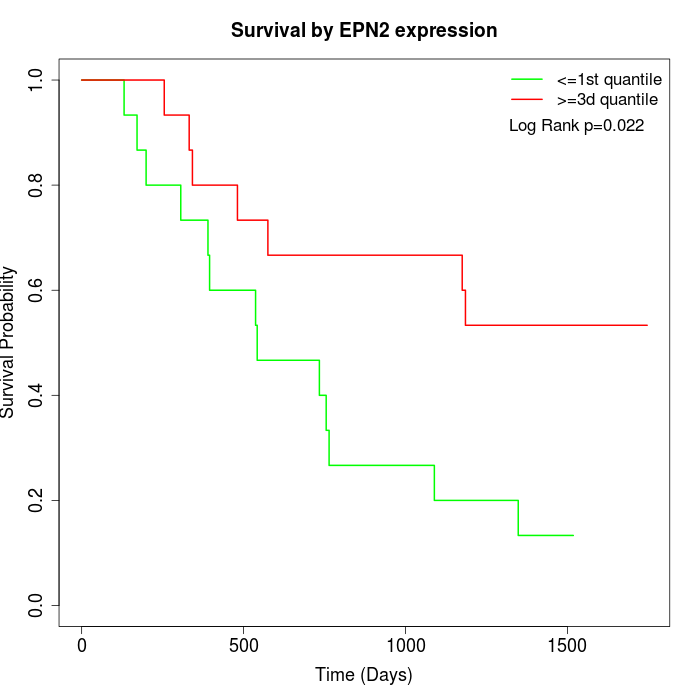
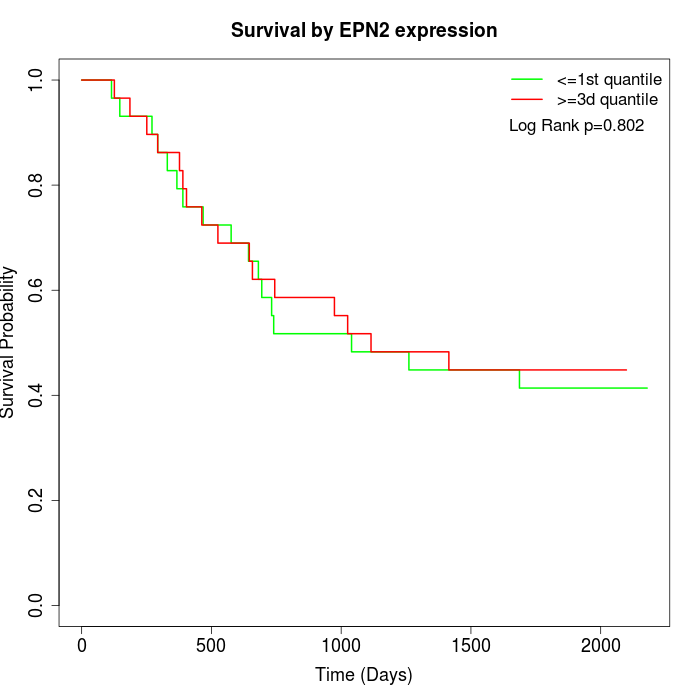
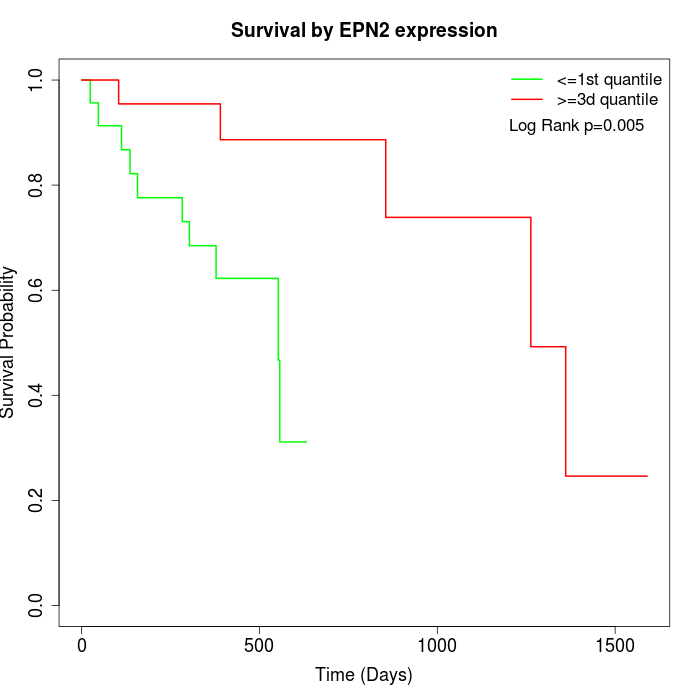
Survival by EPN2 expression:
Note: Click image to view full size file.
Copy number change of EPN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPN2 | 22905 | 4 | 3 | 23 | |
| GSE20123 | EPN2 | 22905 | 4 | 4 | 22 | |
| GSE43470 | EPN2 | 22905 | 1 | 5 | 37 | |
| GSE46452 | EPN2 | 22905 | 34 | 1 | 24 | |
| GSE47630 | EPN2 | 22905 | 7 | 1 | 32 | |
| GSE54993 | EPN2 | 22905 | 3 | 3 | 64 | |
| GSE54994 | EPN2 | 22905 | 6 | 6 | 41 | |
| GSE60625 | EPN2 | 22905 | 4 | 0 | 7 | |
| GSE74703 | EPN2 | 22905 | 1 | 2 | 33 | |
| GSE74704 | EPN2 | 22905 | 3 | 1 | 16 | |
| TCGA | EPN2 | 22905 | 19 | 24 | 53 |
Total number of gains: 86; Total number of losses: 50; Total Number of normals: 352.
Somatic mutations of EPN2:
Generating mutation plots.
Highly correlated genes for EPN2:
Showing top 20/37 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPN2 | SLC48A1 | 0.670634 | 3 | 0 | 3 |
| EPN2 | ADAMTSL4 | 0.662699 | 3 | 0 | 3 |
| EPN2 | NDRG2 | 0.654769 | 3 | 0 | 3 |
| EPN2 | MAST4 | 0.641646 | 3 | 0 | 3 |
| EPN2 | ARL4D | 0.633787 | 3 | 0 | 3 |
| EPN2 | TIMM8B | 0.632484 | 3 | 0 | 3 |
| EPN2 | ERBB2 | 0.626231 | 3 | 0 | 3 |
| EPN2 | MAFG | 0.624321 | 3 | 0 | 3 |
| EPN2 | DKK4 | 0.623501 | 3 | 0 | 3 |
| EPN2 | ALS2CL | 0.620508 | 3 | 0 | 3 |
| EPN2 | SNX21 | 0.616533 | 3 | 0 | 3 |
| EPN2 | ACAP3 | 0.61099 | 3 | 0 | 3 |
| EPN2 | PCDH1 | 0.603557 | 3 | 0 | 3 |
| EPN2 | PHLDB3 | 0.603495 | 4 | 0 | 3 |
| EPN2 | MAPT | 0.602732 | 3 | 0 | 3 |
| EPN2 | ARHGAP10 | 0.601922 | 3 | 0 | 3 |
| EPN2 | AHNAK | 0.601358 | 3 | 0 | 3 |
| EPN2 | BDH1 | 0.599078 | 3 | 0 | 3 |
| EPN2 | TP53INP2 | 0.598199 | 3 | 0 | 3 |
| EPN2 | BLVRB | 0.590472 | 3 | 0 | 3 |
For details and further investigation, click here