| Full name: microtubule associated protein tau | Alias Symbol: MTBT1|tau|PPND|FTDP-17|TAU|MSTD|MTBT2|FLJ31424|MGC138549|PPP1R103 | ||
| Type: protein-coding gene | Cytoband: 17q21.31 | ||
| Entrez ID: 4137 | HGNC ID: HGNC:6893 | Ensembl Gene: ENSG00000186868 | OMIM ID: 157140 |
| Drug and gene relationship at DGIdb | |||
MAPT involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa05010 | Alzheimer's disease |
Expression of MAPT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAPT | 4137 | 203928_x_at | -1.1982 | 0.0476 | |
| GSE20347 | MAPT | 4137 | 203928_x_at | -0.9151 | 0.0000 | |
| GSE23400 | MAPT | 4137 | 203928_x_at | -0.3356 | 0.0000 | |
| GSE26886 | MAPT | 4137 | 203928_x_at | -1.0863 | 0.0000 | |
| GSE29001 | MAPT | 4137 | 203928_x_at | -0.8379 | 0.0000 | |
| GSE38129 | MAPT | 4137 | 203928_x_at | -0.9387 | 0.0000 | |
| GSE45670 | MAPT | 4137 | 203928_x_at | -1.1828 | 0.0000 | |
| GSE53622 | MAPT | 4137 | 59910 | -2.1931 | 0.0000 | |
| GSE53624 | MAPT | 4137 | 59910 | -1.9893 | 0.0000 | |
| GSE63941 | MAPT | 4137 | 203928_x_at | 0.8086 | 0.0158 | |
| GSE77861 | MAPT | 4137 | 225379_at | -0.0793 | 0.5904 | |
| GSE97050 | MAPT | A_21_P0014031 | -0.9215 | 0.1523 | ||
| SRP007169 | MAPT | 4137 | RNAseq | -2.7909 | 0.0000 | |
| SRP064894 | MAPT | 4137 | RNAseq | -2.6777 | 0.0000 | |
| SRP133303 | MAPT | 4137 | RNAseq | -2.9186 | 0.0000 | |
| SRP159526 | MAPT | 4137 | RNAseq | -3.7341 | 0.0000 | |
| SRP193095 | MAPT | 4137 | RNAseq | -1.7779 | 0.0000 | |
| SRP219564 | MAPT | 4137 | RNAseq | -1.7052 | 0.0187 | |
| TCGA | MAPT | 4137 | RNAseq | -0.9584 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 11.
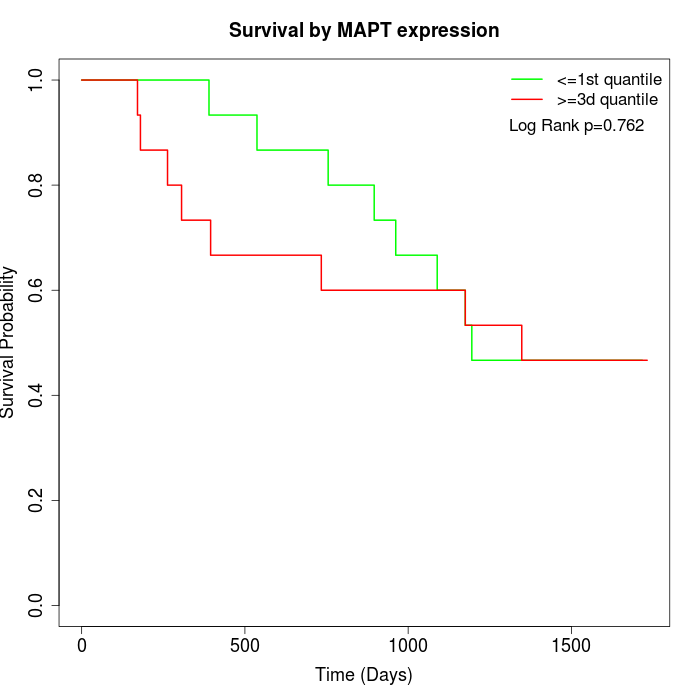
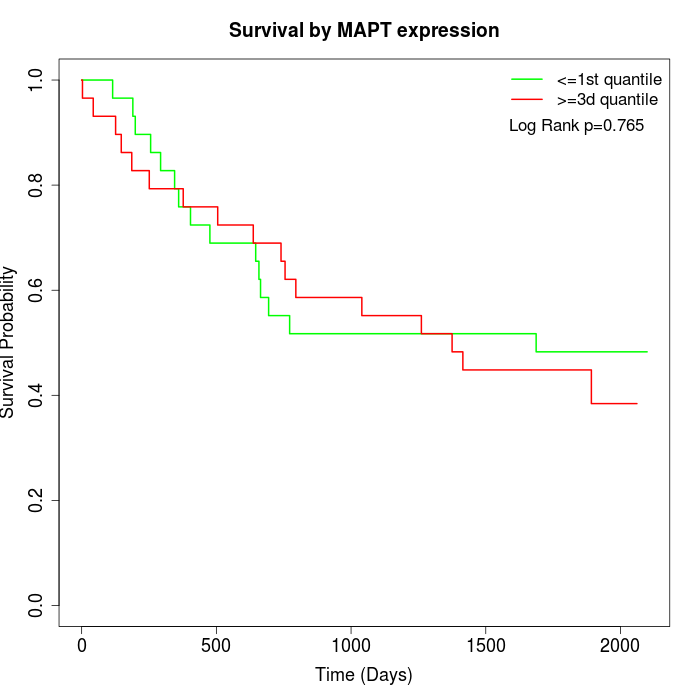
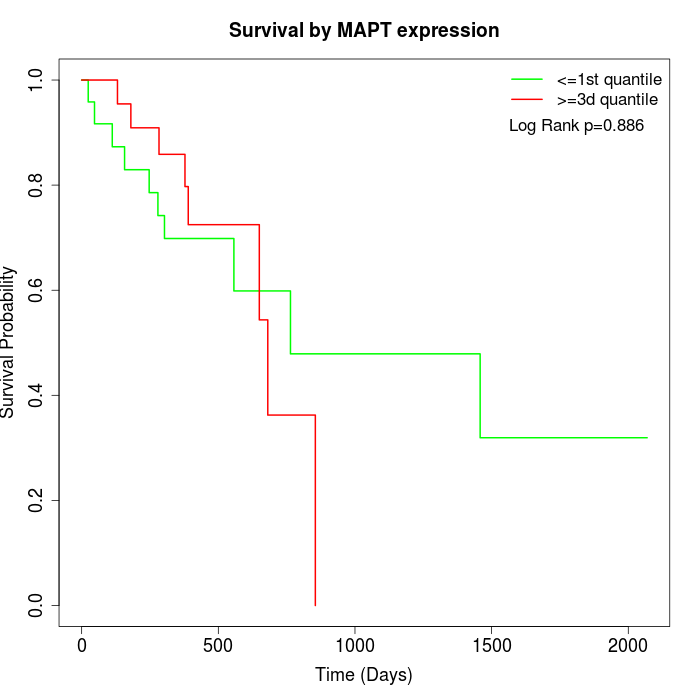
Survival by MAPT expression:
Note: Click image to view full size file.
Copy number change of MAPT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAPT | 4137 | 6 | 2 | 22 | |
| GSE20123 | MAPT | 4137 | 6 | 2 | 22 | |
| GSE43470 | MAPT | 4137 | 1 | 2 | 40 | |
| GSE46452 | MAPT | 4137 | 32 | 0 | 27 | |
| GSE47630 | MAPT | 4137 | 9 | 0 | 31 | |
| GSE54993 | MAPT | 4137 | 3 | 4 | 63 | |
| GSE54994 | MAPT | 4137 | 9 | 6 | 38 | |
| GSE60625 | MAPT | 4137 | 4 | 0 | 7 | |
| GSE74703 | MAPT | 4137 | 1 | 1 | 34 | |
| GSE74704 | MAPT | 4137 | 4 | 1 | 15 | |
| TCGA | MAPT | 4137 | 26 | 7 | 63 |
Total number of gains: 101; Total number of losses: 25; Total Number of normals: 362.
Somatic mutations of MAPT:
Generating mutation plots.
Highly correlated genes for MAPT:
Showing top 20/934 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAPT | MYZAP | 0.821621 | 3 | 0 | 3 |
| MAPT | EDN3 | 0.760641 | 10 | 0 | 10 |
| MAPT | ENDOU | 0.758139 | 9 | 0 | 9 |
| MAPT | HCG22 | 0.752759 | 6 | 0 | 6 |
| MAPT | NUCB2 | 0.749571 | 9 | 0 | 9 |
| MAPT | GPX6 | 0.748443 | 3 | 0 | 3 |
| MAPT | SFTA2 | 0.747641 | 3 | 0 | 3 |
| MAPT | SAMD5 | 0.747253 | 5 | 0 | 5 |
| MAPT | TFAP2B | 0.745944 | 9 | 0 | 9 |
| MAPT | SLURP1 | 0.740221 | 10 | 0 | 10 |
| MAPT | SLC16A6 | 0.739995 | 9 | 0 | 9 |
| MAPT | PAQR8 | 0.737745 | 5 | 0 | 5 |
| MAPT | BNIPL | 0.737511 | 5 | 0 | 5 |
| MAPT | GPD1L | 0.732164 | 9 | 0 | 9 |
| MAPT | GYS2 | 0.731798 | 9 | 0 | 9 |
| MAPT | ZNF662 | 0.727669 | 5 | 0 | 5 |
| MAPT | UPK1A | 0.722675 | 11 | 0 | 10 |
| MAPT | SPINK7 | 0.721451 | 6 | 0 | 6 |
| MAPT | CRISP3 | 0.72082 | 10 | 0 | 9 |
| MAPT | GDPD3 | 0.715491 | 10 | 0 | 9 |
For details and further investigation, click here