| Full name: ERCC excision repair 6 like, spindle assembly checkpoint helicase | Alias Symbol: FLJ20105|PICH|RAD26L | ||
| Type: protein-coding gene | Cytoband: Xq13.1 | ||
| Entrez ID: 54821 | HGNC ID: HGNC:20794 | Ensembl Gene: ENSG00000186871 | OMIM ID: 300687 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ERCC6L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ERCC6L | 54821 | 219650_at | 0.8730 | 0.0339 | |
| GSE20347 | ERCC6L | 54821 | 219650_at | 0.6318 | 0.0000 | |
| GSE23400 | ERCC6L | 54821 | 219650_at | 0.2924 | 0.0000 | |
| GSE26886 | ERCC6L | 54821 | 219650_at | 0.1711 | 0.4587 | |
| GSE29001 | ERCC6L | 54821 | 219650_at | 0.8420 | 0.0017 | |
| GSE38129 | ERCC6L | 54821 | 219650_at | 0.9377 | 0.0000 | |
| GSE45670 | ERCC6L | 54821 | 219650_at | 0.8098 | 0.0000 | |
| GSE53622 | ERCC6L | 54821 | 91293 | 1.5149 | 0.0000 | |
| GSE53624 | ERCC6L | 54821 | 91293 | 1.6702 | 0.0000 | |
| GSE63941 | ERCC6L | 54821 | 219650_at | 1.6025 | 0.0129 | |
| GSE77861 | ERCC6L | 54821 | 219650_at | 0.2503 | 0.1504 | |
| GSE97050 | ERCC6L | 54821 | A_23_P96325 | 0.1410 | 0.8758 | |
| SRP007169 | ERCC6L | 54821 | RNAseq | 1.7577 | 0.0019 | |
| SRP008496 | ERCC6L | 54821 | RNAseq | 1.2397 | 0.0101 | |
| SRP064894 | ERCC6L | 54821 | RNAseq | 1.4268 | 0.0000 | |
| SRP133303 | ERCC6L | 54821 | RNAseq | 1.0751 | 0.0000 | |
| SRP159526 | ERCC6L | 54821 | RNAseq | 0.7972 | 0.0103 | |
| SRP193095 | ERCC6L | 54821 | RNAseq | 0.7466 | 0.0000 | |
| SRP219564 | ERCC6L | 54821 | RNAseq | 1.0020 | 0.1071 | |
| TCGA | ERCC6L | 54821 | RNAseq | 1.0942 | 0.0000 |
Upregulated datasets: 8; Downregulated datasets: 0.
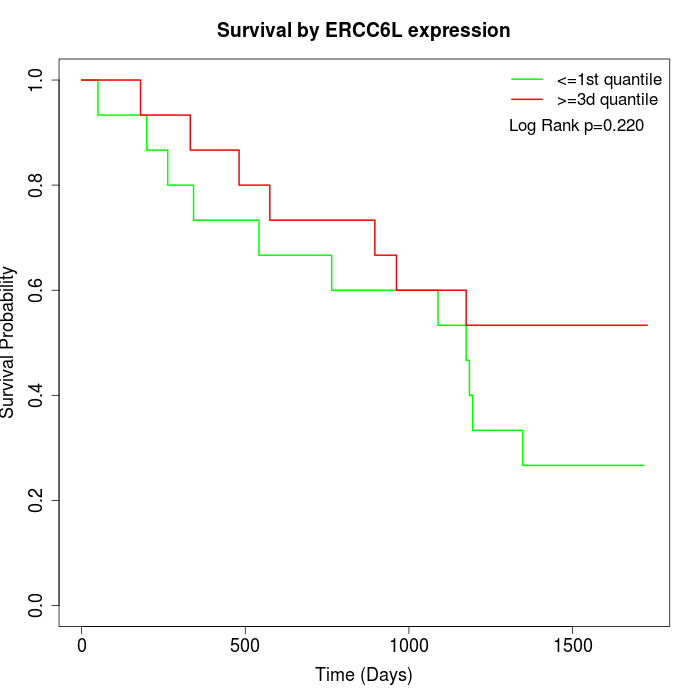
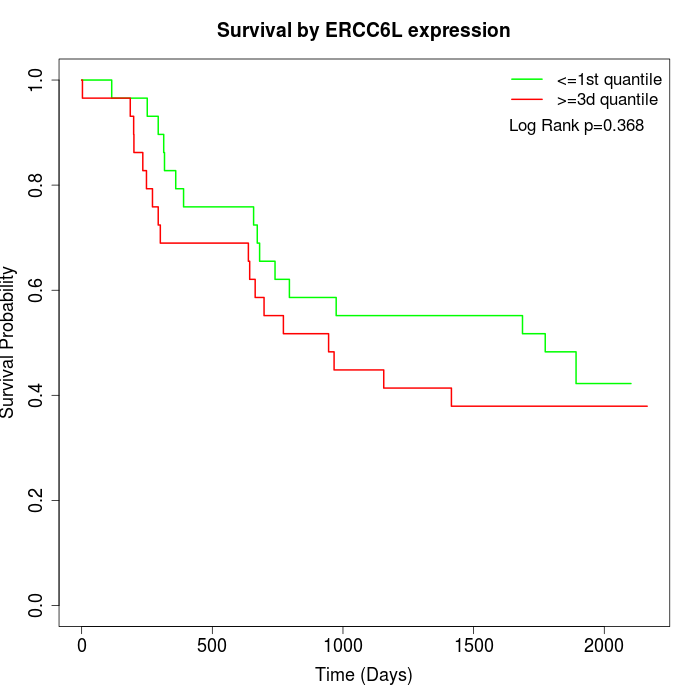
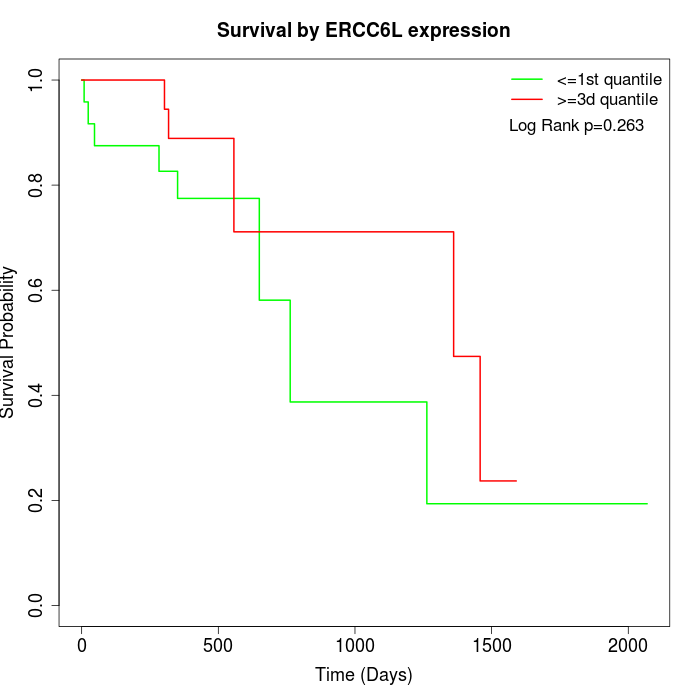
Survival by ERCC6L expression:
Note: Click image to view full size file.
Copy number change of ERCC6L:
No record found for this gene.
Somatic mutations of ERCC6L:
Generating mutation plots.
Highly correlated genes for ERCC6L:
Showing top 20/1572 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ERCC6L | CDC6 | 0.791632 | 9 | 0 | 9 |
| ERCC6L | DEPDC1B | 0.78799 | 6 | 0 | 6 |
| ERCC6L | STIL | 0.786208 | 10 | 0 | 10 |
| ERCC6L | CDC20 | 0.781765 | 10 | 0 | 10 |
| ERCC6L | KIF4A | 0.781364 | 11 | 0 | 10 |
| ERCC6L | CDCA3 | 0.780319 | 11 | 0 | 10 |
| ERCC6L | C1orf112 | 0.776448 | 9 | 0 | 8 |
| ERCC6L | GINS1 | 0.774763 | 10 | 0 | 10 |
| ERCC6L | EXO1 | 0.773308 | 11 | 0 | 11 |
| ERCC6L | MCM6 | 0.771985 | 10 | 0 | 10 |
| ERCC6L | RFC4 | 0.767624 | 10 | 0 | 10 |
| ERCC6L | CEP55 | 0.766572 | 11 | 0 | 10 |
| ERCC6L | GMNN | 0.76495 | 11 | 0 | 10 |
| ERCC6L | KIF18A | 0.763188 | 11 | 0 | 10 |
| ERCC6L | BUB1 | 0.762897 | 9 | 0 | 8 |
| ERCC6L | MCM10 | 0.760572 | 10 | 0 | 10 |
| ERCC6L | TOP2A | 0.758621 | 10 | 0 | 10 |
| ERCC6L | MELK | 0.756628 | 11 | 0 | 10 |
| ERCC6L | FBXO5 | 0.756586 | 10 | 0 | 10 |
| ERCC6L | RAD51 | 0.756132 | 11 | 0 | 10 |
For details and further investigation, click here