| Full name: growth arrest and DNA damage inducible beta | Alias Symbol: GADD45BETA|DKFZP566B133 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 4616 | HGNC ID: HGNC:4096 | Ensembl Gene: ENSG00000099860 | OMIM ID: 604948 |
| Drug and gene relationship at DGIdb | |||
GADD45B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04068 | FoxO signaling pathway | |
| hsa04110 | Cell cycle | |
| hsa04115 | p53 signaling pathway | |
| hsa04210 | Apoptosis |
Expression of GADD45B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GADD45B | 4616 | 207574_s_at | -1.2132 | 0.4626 | |
| GSE20347 | GADD45B | 4616 | 207574_s_at | -0.6053 | 0.0038 | |
| GSE23400 | GADD45B | 4616 | 209304_x_at | -0.8484 | 0.0000 | |
| GSE26886 | GADD45B | 4616 | 207574_s_at | 0.0085 | 0.9870 | |
| GSE29001 | GADD45B | 4616 | 207574_s_at | -1.3516 | 0.0063 | |
| GSE38129 | GADD45B | 4616 | 207574_s_at | -1.0492 | 0.0002 | |
| GSE45670 | GADD45B | 4616 | 207574_s_at | -1.4735 | 0.0000 | |
| GSE53622 | GADD45B | 4616 | 114099 | -0.9723 | 0.0000 | |
| GSE53624 | GADD45B | 4616 | 11902 | -1.0951 | 0.0000 | |
| GSE63941 | GADD45B | 4616 | 207574_s_at | -1.6638 | 0.0808 | |
| GSE77861 | GADD45B | 4616 | 209304_x_at | -0.3055 | 0.2665 | |
| GSE97050 | GADD45B | 4616 | A_24_P239606 | -0.7125 | 0.2882 | |
| SRP007169 | GADD45B | 4616 | RNAseq | -2.9226 | 0.0000 | |
| SRP008496 | GADD45B | 4616 | RNAseq | -2.0193 | 0.0000 | |
| SRP064894 | GADD45B | 4616 | RNAseq | -0.6594 | 0.0971 | |
| SRP133303 | GADD45B | 4616 | RNAseq | -0.4114 | 0.2248 | |
| SRP159526 | GADD45B | 4616 | RNAseq | -0.6019 | 0.2721 | |
| SRP193095 | GADD45B | 4616 | RNAseq | -0.8124 | 0.0019 | |
| SRP219564 | GADD45B | 4616 | RNAseq | -0.3536 | 0.6986 | |
| TCGA | GADD45B | 4616 | RNAseq | -0.5631 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 6.
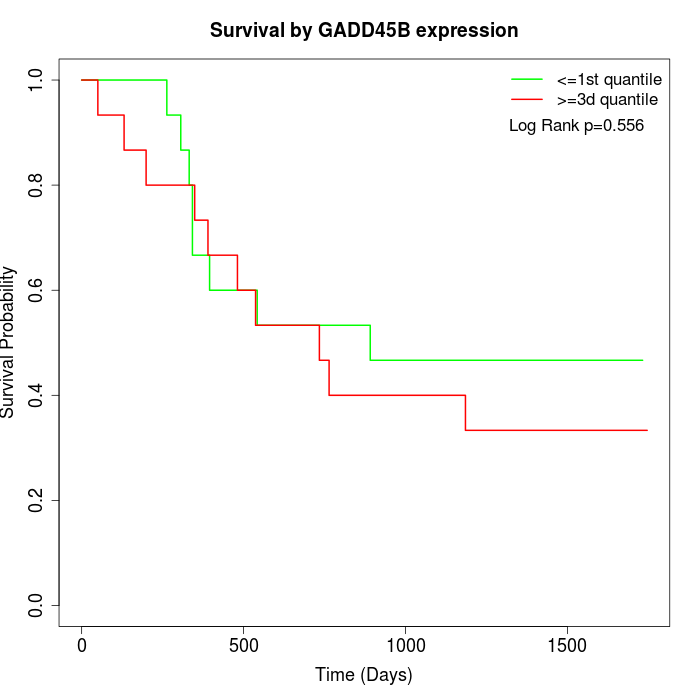
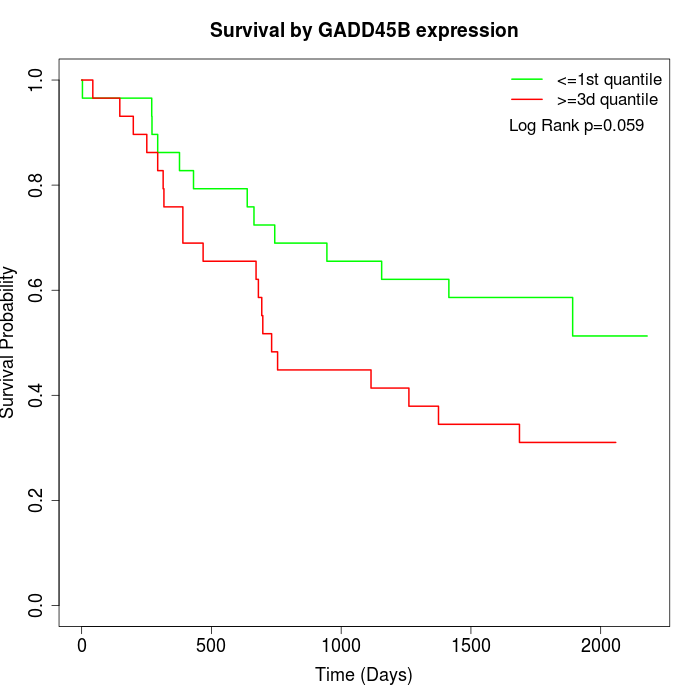
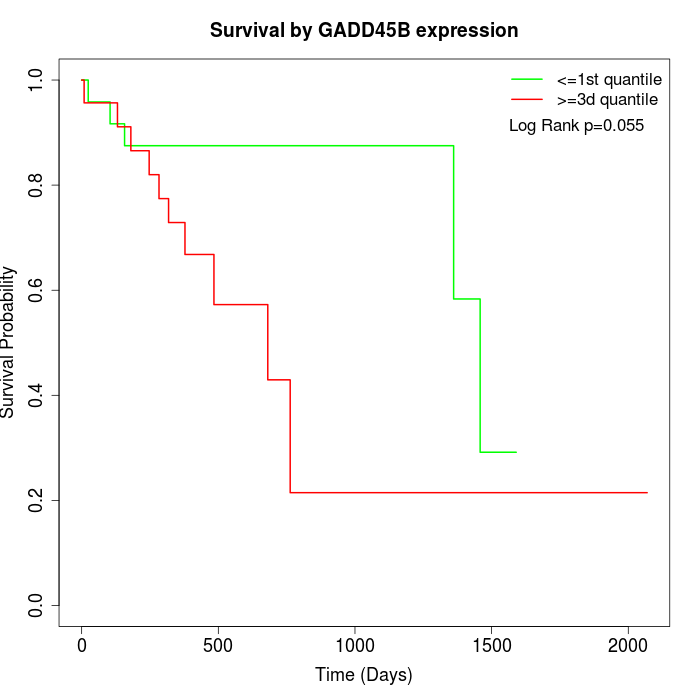
Survival by GADD45B expression:
Note: Click image to view full size file.
Copy number change of GADD45B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GADD45B | 4616 | 4 | 4 | 22 | |
| GSE20123 | GADD45B | 4616 | 4 | 4 | 22 | |
| GSE43470 | GADD45B | 4616 | 1 | 10 | 32 | |
| GSE46452 | GADD45B | 4616 | 47 | 1 | 11 | |
| GSE47630 | GADD45B | 4616 | 5 | 7 | 28 | |
| GSE54993 | GADD45B | 4616 | 16 | 3 | 51 | |
| GSE54994 | GADD45B | 4616 | 8 | 15 | 30 | |
| GSE60625 | GADD45B | 4616 | 9 | 0 | 2 | |
| GSE74703 | GADD45B | 4616 | 1 | 7 | 28 | |
| GSE74704 | GADD45B | 4616 | 2 | 3 | 15 | |
| TCGA | GADD45B | 4616 | 6 | 24 | 66 |
Total number of gains: 103; Total number of losses: 78; Total Number of normals: 307.
Somatic mutations of GADD45B:
Generating mutation plots.
Highly correlated genes for GADD45B:
Showing top 20/644 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GADD45B | CSRNP1 | 0.753235 | 8 | 0 | 8 |
| GADD45B | SORCS1 | 0.726096 | 3 | 0 | 3 |
| GADD45B | CYP21A2 | 0.710924 | 3 | 0 | 3 |
| GADD45B | SIN3A | 0.698904 | 3 | 0 | 3 |
| GADD45B | SLC25A25 | 0.696154 | 4 | 0 | 3 |
| GADD45B | CLEC3B | 0.695043 | 3 | 0 | 3 |
| GADD45B | TBC1D20 | 0.693264 | 3 | 0 | 3 |
| GADD45B | NR4A1 | 0.687749 | 6 | 0 | 5 |
| GADD45B | SKI | 0.681671 | 3 | 0 | 3 |
| GADD45B | CFL2 | 0.679793 | 4 | 0 | 4 |
| GADD45B | RBPMS2 | 0.677576 | 4 | 0 | 4 |
| GADD45B | PDCD4 | 0.677216 | 4 | 0 | 4 |
| GADD45B | HMCN1 | 0.675142 | 3 | 0 | 3 |
| GADD45B | RGS3 | 0.675104 | 3 | 0 | 3 |
| GADD45B | RGS2 | 0.670001 | 8 | 0 | 8 |
| GADD45B | HSPB7 | 0.669656 | 8 | 0 | 8 |
| GADD45B | SERTAD1 | 0.661355 | 6 | 0 | 5 |
| GADD45B | DUSP1 | 0.658157 | 12 | 0 | 10 |
| GADD45B | AOC3 | 0.657394 | 9 | 0 | 8 |
| GADD45B | SLMAP | 0.656758 | 5 | 0 | 4 |
For details and further investigation, click here