| Full name: katanin regulatory subunit B1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16q21 | ||
| Entrez ID: 10300 | HGNC ID: HGNC:6217 | Ensembl Gene: ENSG00000140854 | OMIM ID: 602703 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of KATNB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KATNB1 | 10300 | 203162_s_at | 0.6304 | 0.0909 | |
| GSE20347 | KATNB1 | 10300 | 203162_s_at | 0.0667 | 0.6894 | |
| GSE23400 | KATNB1 | 10300 | 203162_s_at | 0.1127 | 0.0347 | |
| GSE26886 | KATNB1 | 10300 | 203162_s_at | 0.2957 | 0.1328 | |
| GSE29001 | KATNB1 | 10300 | 203162_s_at | 0.0633 | 0.8663 | |
| GSE38129 | KATNB1 | 10300 | 203162_s_at | 0.2317 | 0.0768 | |
| GSE45670 | KATNB1 | 10300 | 203162_s_at | 0.2241 | 0.0291 | |
| GSE53622 | KATNB1 | 10300 | 93522 | 0.1312 | 0.1969 | |
| GSE53624 | KATNB1 | 10300 | 93522 | 0.2949 | 0.0000 | |
| GSE63941 | KATNB1 | 10300 | 203162_s_at | -0.0723 | 0.8799 | |
| GSE77861 | KATNB1 | 10300 | 203162_s_at | 0.1704 | 0.4932 | |
| GSE97050 | KATNB1 | 10300 | A_33_P3396129 | -0.1553 | 0.6110 | |
| SRP007169 | KATNB1 | 10300 | RNAseq | -0.1903 | 0.6558 | |
| SRP008496 | KATNB1 | 10300 | RNAseq | -0.1830 | 0.5677 | |
| SRP064894 | KATNB1 | 10300 | RNAseq | 0.0944 | 0.6021 | |
| SRP133303 | KATNB1 | 10300 | RNAseq | 0.3320 | 0.1293 | |
| SRP159526 | KATNB1 | 10300 | RNAseq | 0.2804 | 0.3208 | |
| SRP193095 | KATNB1 | 10300 | RNAseq | 0.3287 | 0.0044 | |
| SRP219564 | KATNB1 | 10300 | RNAseq | -0.1212 | 0.7504 | |
| TCGA | KATNB1 | 10300 | RNAseq | 0.2393 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
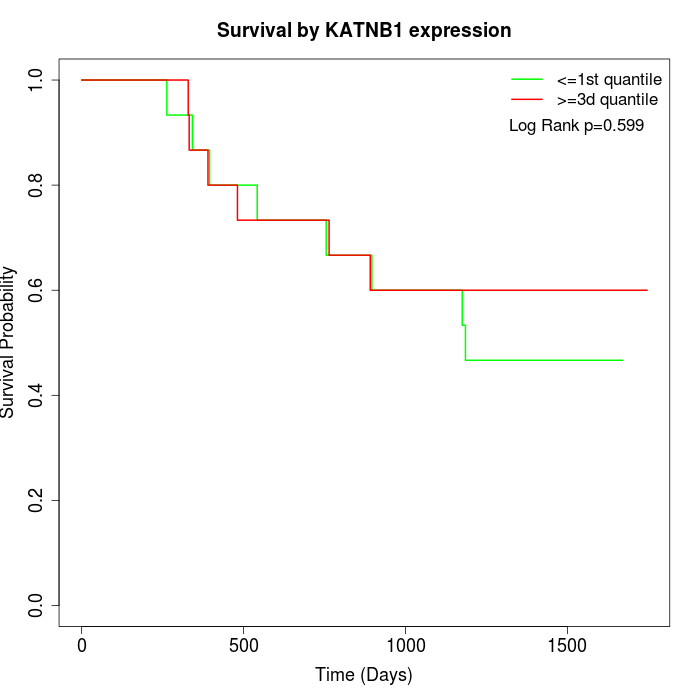
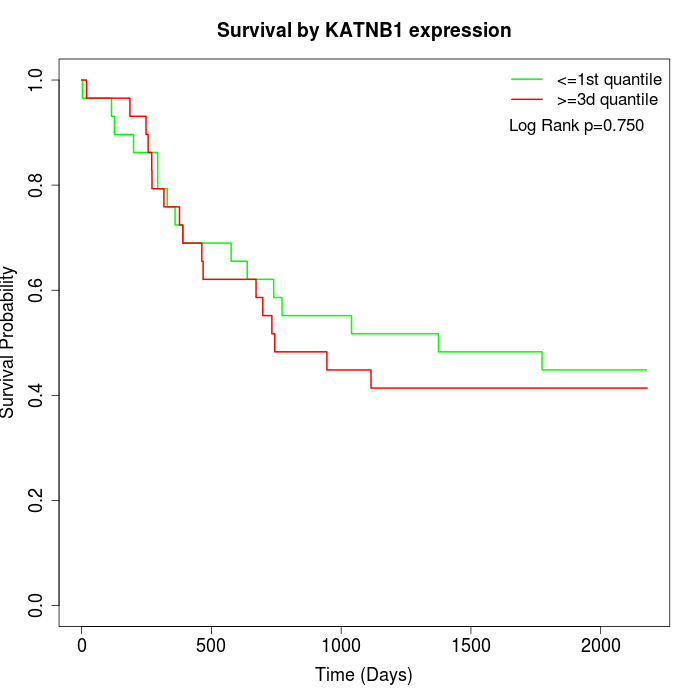
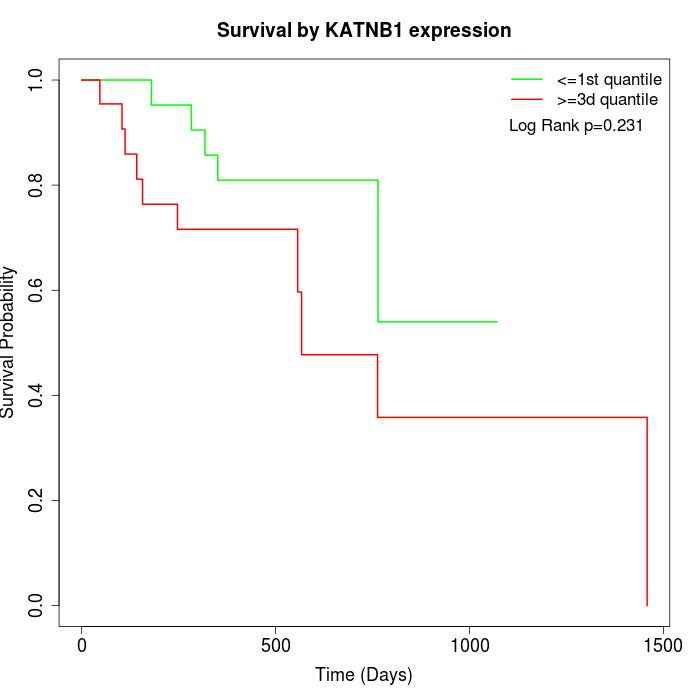
Survival by KATNB1 expression:
Note: Click image to view full size file.
Copy number change of KATNB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KATNB1 | 10300 | 6 | 0 | 24 | |
| GSE20123 | KATNB1 | 10300 | 6 | 1 | 23 | |
| GSE43470 | KATNB1 | 10300 | 2 | 6 | 35 | |
| GSE46452 | KATNB1 | 10300 | 38 | 1 | 20 | |
| GSE47630 | KATNB1 | 10300 | 10 | 8 | 22 | |
| GSE54993 | KATNB1 | 10300 | 2 | 4 | 64 | |
| GSE54994 | KATNB1 | 10300 | 6 | 10 | 37 | |
| GSE60625 | KATNB1 | 10300 | 4 | 0 | 7 | |
| GSE74703 | KATNB1 | 10300 | 2 | 4 | 30 | |
| GSE74704 | KATNB1 | 10300 | 5 | 0 | 15 | |
| TCGA | KATNB1 | 10300 | 26 | 12 | 58 |
Total number of gains: 107; Total number of losses: 46; Total Number of normals: 335.
Somatic mutations of KATNB1:
Generating mutation plots.
Highly correlated genes for KATNB1:
Showing top 20/248 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KATNB1 | TRAF7 | 0.709853 | 4 | 0 | 4 |
| KATNB1 | MCCC2 | 0.660189 | 3 | 0 | 3 |
| KATNB1 | EMC8 | 0.640634 | 7 | 0 | 7 |
| KATNB1 | MTCH2 | 0.63852 | 3 | 0 | 3 |
| KATNB1 | AGTRAP | 0.634724 | 4 | 0 | 3 |
| KATNB1 | DNTTIP1 | 0.628161 | 5 | 0 | 3 |
| KATNB1 | TMF1 | 0.627069 | 3 | 0 | 3 |
| KATNB1 | CHPF | 0.624841 | 6 | 0 | 5 |
| KATNB1 | MRPL12 | 0.624555 | 3 | 0 | 3 |
| KATNB1 | BCAR1 | 0.622785 | 4 | 0 | 3 |
| KATNB1 | RALY-AS1 | 0.617561 | 3 | 0 | 3 |
| KATNB1 | PTRH1 | 0.61698 | 5 | 0 | 4 |
| KATNB1 | SPNS1 | 0.616625 | 4 | 0 | 3 |
| KATNB1 | MRPS30 | 0.61413 | 3 | 0 | 3 |
| KATNB1 | ZC3HC1 | 0.611423 | 4 | 0 | 3 |
| KATNB1 | GINS3 | 0.611161 | 8 | 0 | 7 |
| KATNB1 | VPS35 | 0.611031 | 5 | 0 | 3 |
| KATNB1 | GP5 | 0.606746 | 3 | 0 | 3 |
| KATNB1 | MRPL37 | 0.606452 | 4 | 0 | 3 |
| KATNB1 | NPM3 | 0.605903 | 5 | 0 | 3 |
For details and further investigation, click here