| Full name: kinase suppressor of ras 2 | Alias Symbol: FLJ25965 | ||
| Type: protein-coding gene | Cytoband: 12q24.22-q24.23 | ||
| Entrez ID: 283455 | HGNC ID: HGNC:18610 | Ensembl Gene: ENSG00000171435 | OMIM ID: 610737 |
| Drug and gene relationship at DGIdb | |||
KSR2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway |
Expression of KSR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KSR2 | 283455 | 1553491_at | 0.2498 | 0.3672 | |
| GSE26886 | KSR2 | 283455 | 1553491_at | 0.1558 | 0.1070 | |
| GSE45670 | KSR2 | 283455 | 1553491_at | 0.1131 | 0.3252 | |
| GSE53622 | KSR2 | 283455 | 26614 | 0.3154 | 0.1217 | |
| GSE53624 | KSR2 | 283455 | 26614 | 0.1128 | 0.5308 | |
| GSE63941 | KSR2 | 283455 | 230551_at | 0.5799 | 0.2006 | |
| GSE77861 | KSR2 | 283455 | 230551_at | 0.0813 | 0.7132 | |
| SRP133303 | KSR2 | 283455 | RNAseq | 0.1915 | 0.4118 | |
| SRP159526 | KSR2 | 283455 | RNAseq | 0.8575 | 0.1101 | |
| SRP193095 | KSR2 | 283455 | RNAseq | 0.2463 | 0.2182 | |
| TCGA | KSR2 | 283455 | RNAseq | -0.8261 | 0.0197 |
Upregulated datasets: 0; Downregulated datasets: 0.
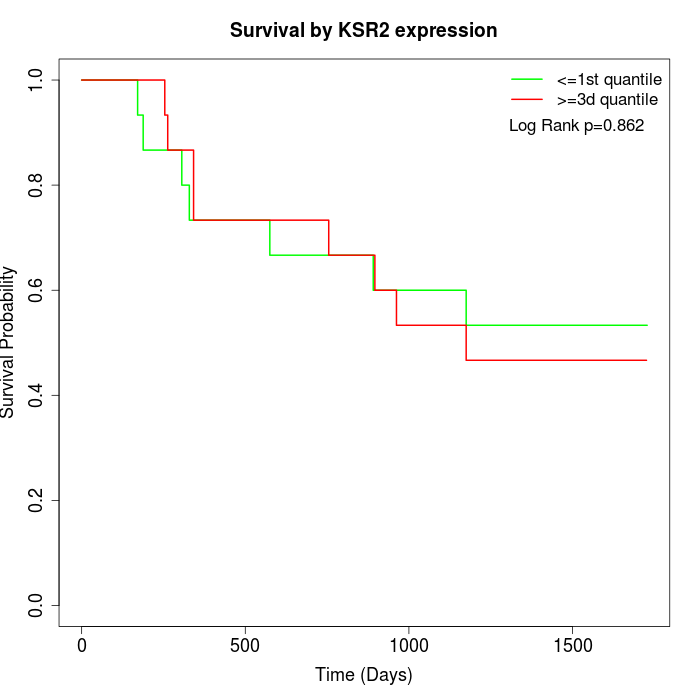
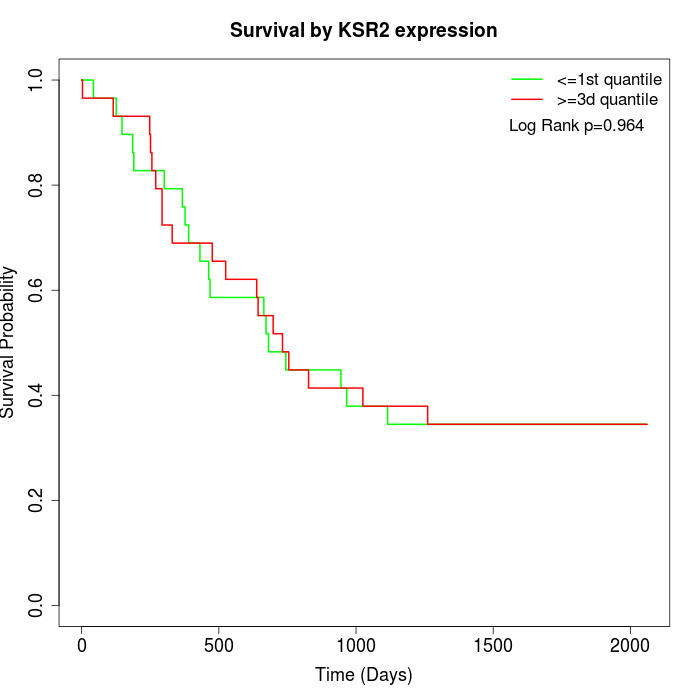
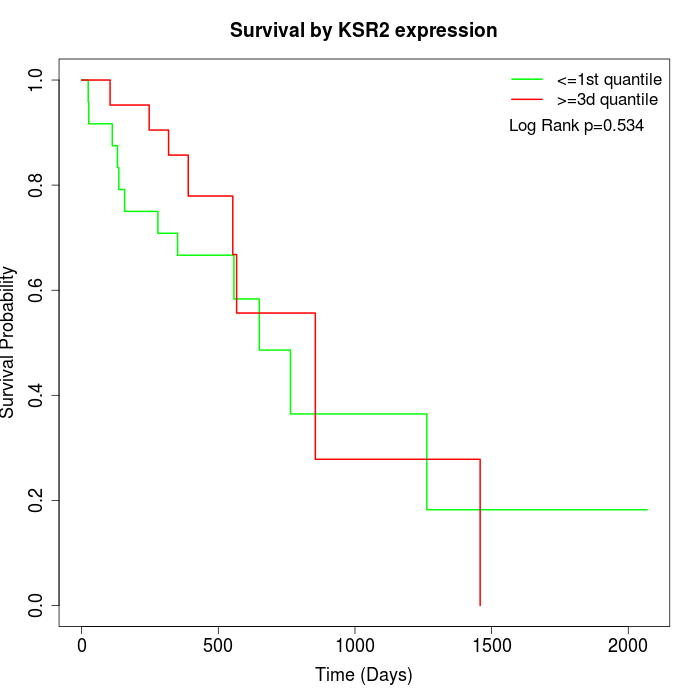
Survival by KSR2 expression:
Note: Click image to view full size file.
Copy number change of KSR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KSR2 | 283455 | 6 | 2 | 22 | |
| GSE20123 | KSR2 | 283455 | 6 | 2 | 22 | |
| GSE43470 | KSR2 | 283455 | 2 | 0 | 41 | |
| GSE46452 | KSR2 | 283455 | 9 | 1 | 49 | |
| GSE47630 | KSR2 | 283455 | 9 | 3 | 28 | |
| GSE54993 | KSR2 | 283455 | 0 | 5 | 65 | |
| GSE54994 | KSR2 | 283455 | 6 | 6 | 41 | |
| GSE60625 | KSR2 | 283455 | 0 | 0 | 11 | |
| GSE74703 | KSR2 | 283455 | 2 | 0 | 34 | |
| GSE74704 | KSR2 | 283455 | 3 | 2 | 15 | |
| TCGA | KSR2 | 283455 | 21 | 11 | 64 |
Total number of gains: 64; Total number of losses: 32; Total Number of normals: 392.
Somatic mutations of KSR2:
Generating mutation plots.
Highly correlated genes for KSR2:
Showing top 20/43 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KSR2 | G0S2 | 0.710671 | 3 | 0 | 3 |
| KSR2 | GPR153 | 0.656239 | 3 | 0 | 3 |
| KSR2 | GRAMD4 | 0.65393 | 3 | 0 | 3 |
| KSR2 | RGS16 | 0.641164 | 3 | 0 | 3 |
| KSR2 | RFPL2 | 0.634332 | 3 | 0 | 3 |
| KSR2 | CPA6 | 0.633778 | 3 | 0 | 3 |
| KSR2 | APBA3 | 0.62487 | 3 | 0 | 3 |
| KSR2 | NGFR | 0.62323 | 3 | 0 | 3 |
| KSR2 | RIMS2 | 0.615222 | 4 | 0 | 3 |
| KSR2 | CFC1 | 0.612361 | 3 | 0 | 3 |
| KSR2 | GRM4 | 0.60605 | 3 | 0 | 3 |
| KSR2 | BCL2A1 | 0.605911 | 3 | 0 | 3 |
| KSR2 | ADD2 | 0.605851 | 3 | 0 | 3 |
| KSR2 | ACTN3 | 0.604332 | 3 | 0 | 3 |
| KSR2 | C20orf141 | 0.602684 | 3 | 0 | 3 |
| KSR2 | TRABD | 0.600639 | 3 | 0 | 3 |
| KSR2 | SCN1B | 0.598256 | 3 | 0 | 3 |
| KSR2 | HSF1 | 0.593409 | 3 | 0 | 3 |
| KSR2 | WDR62 | 0.588836 | 3 | 0 | 3 |
| KSR2 | KIF1A | 0.588683 | 4 | 0 | 3 |
For details and further investigation, click here