| Full name: long intergenic non-protein coding RNA 1233 | Alias Symbol: XLOC_013014 | ||
| Type: non-coding RNA | Cytoband: 19p12 | ||
| Entrez ID: 100128139 | HGNC ID: HGNC:49756 | Ensembl Gene: ENSG00000269364 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC01233:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC01233 | 100128139 | 236768_at | -0.0405 | 0.8573 | |
| GSE26886 | LINC01233 | 100128139 | 236768_at | 0.0472 | 0.6099 | |
| GSE45670 | LINC01233 | 100128139 | 236768_at | 0.0106 | 0.8837 | |
| GSE53622 | LINC01233 | 100128139 | 23234 | 0.0485 | 0.8515 | |
| GSE53624 | LINC01233 | 100128139 | 23234 | 0.0450 | 0.7402 | |
| GSE63941 | LINC01233 | 100128139 | 236768_at | -0.0080 | 0.9566 | |
| GSE77861 | LINC01233 | 100128139 | 236768_at | -0.0596 | 0.5416 |
Upregulated datasets: 0; Downregulated datasets: 0.
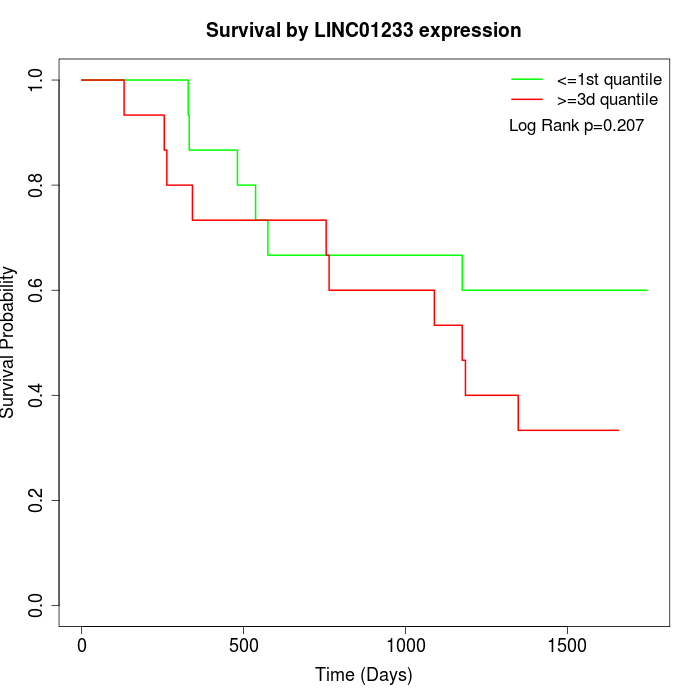
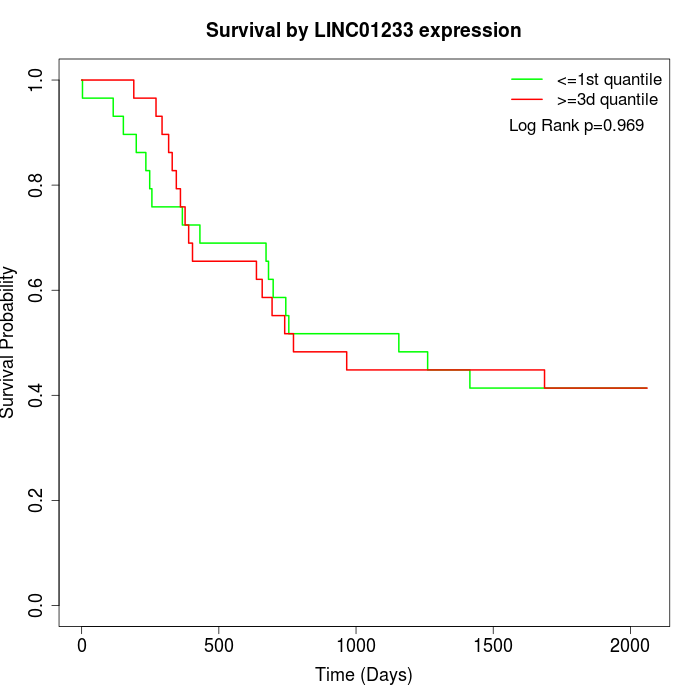
Survival by LINC01233 expression:
Note: Click image to view full size file.
Copy number change of LINC01233:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC01233 | 100128139 | 5 | 4 | 21 | |
| GSE20123 | LINC01233 | 100128139 | 5 | 3 | 22 | |
| GSE43470 | LINC01233 | 100128139 | 2 | 6 | 35 | |
| GSE46452 | LINC01233 | 100128139 | 47 | 1 | 11 | |
| GSE47630 | LINC01233 | 100128139 | 7 | 7 | 26 | |
| GSE54993 | LINC01233 | 100128139 | 17 | 3 | 50 | |
| GSE54994 | LINC01233 | 100128139 | 6 | 13 | 34 | |
| GSE60625 | LINC01233 | 100128139 | 9 | 0 | 2 | |
| GSE74703 | LINC01233 | 100128139 | 2 | 4 | 30 | |
| GSE74704 | LINC01233 | 100128139 | 2 | 3 | 15 |
Total number of gains: 102; Total number of losses: 44; Total Number of normals: 246.
Somatic mutations of LINC01233:
Generating mutation plots.
Highly correlated genes for LINC01233:
Showing top 20/27 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC01233 | MS4A3 | 0.722835 | 3 | 0 | 3 |
| LINC01233 | LILRB5 | 0.649653 | 3 | 0 | 3 |
| LINC01233 | MIA | 0.643634 | 3 | 0 | 3 |
| LINC01233 | DERL3 | 0.635165 | 3 | 0 | 3 |
| LINC01233 | NPPB | 0.608162 | 3 | 0 | 3 |
| LINC01233 | TCAP | 0.60475 | 3 | 0 | 3 |
| LINC01233 | DEFB125 | 0.596609 | 4 | 0 | 3 |
| LINC01233 | CLEC1B | 0.592656 | 4 | 0 | 3 |
| LINC01233 | PRM1 | 0.585605 | 3 | 0 | 3 |
| LINC01233 | ABCC8 | 0.583025 | 3 | 0 | 3 |
| LINC01233 | SPATA25 | 0.580505 | 3 | 0 | 3 |
| LINC01233 | INS | 0.580412 | 4 | 0 | 3 |
| LINC01233 | S100A1 | 0.579264 | 3 | 0 | 3 |
| LINC01233 | HCST | 0.57134 | 3 | 0 | 3 |
| LINC01233 | TSPAN16 | 0.566973 | 4 | 0 | 3 |
| LINC01233 | CCL27 | 0.55672 | 3 | 0 | 3 |
| LINC01233 | GSG1 | 0.555427 | 3 | 0 | 3 |
| LINC01233 | KCNQ4 | 0.55056 | 4 | 0 | 3 |
| LINC01233 | CABP1 | 0.545489 | 3 | 0 | 3 |
| LINC01233 | DLGAP2 | 0.541508 | 4 | 0 | 3 |
For details and further investigation, click here