| Full name: minichromosome maintenance complex component 2 | Alias Symbol: D3S3194|KIAA0030|BM28|cdc19|DFNA70 | ||
| Type: protein-coding gene | Cytoband: 3q21.3 | ||
| Entrez ID: 4171 | HGNC ID: HGNC:6944 | Ensembl Gene: ENSG00000073111 | OMIM ID: 116945 |
| Drug and gene relationship at DGIdb | |||
MCM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle |
Expression of MCM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MCM2 | 4171 | 202107_s_at | 1.7309 | 0.0123 | |
| GSE20347 | MCM2 | 4171 | 202107_s_at | 2.0328 | 0.0000 | |
| GSE23400 | MCM2 | 4171 | 202107_s_at | 1.5433 | 0.0000 | |
| GSE26886 | MCM2 | 4171 | 202107_s_at | 0.8306 | 0.0015 | |
| GSE29001 | MCM2 | 4171 | 202107_s_at | 1.7578 | 0.0001 | |
| GSE38129 | MCM2 | 4171 | 202107_s_at | 2.1310 | 0.0000 | |
| GSE45670 | MCM2 | 4171 | 202107_s_at | 1.4970 | 0.0000 | |
| GSE53622 | MCM2 | 4171 | 4355 | 1.7260 | 0.0000 | |
| GSE53624 | MCM2 | 4171 | 4355 | 1.6447 | 0.0000 | |
| GSE63941 | MCM2 | 4171 | 202107_s_at | 3.8947 | 0.0000 | |
| GSE77861 | MCM2 | 4171 | 202107_s_at | 1.4252 | 0.0007 | |
| GSE97050 | MCM2 | 4171 | A_32_P103633 | 0.8171 | 0.1645 | |
| SRP007169 | MCM2 | 4171 | RNAseq | 2.9021 | 0.0000 | |
| SRP008496 | MCM2 | 4171 | RNAseq | 2.5867 | 0.0000 | |
| SRP064894 | MCM2 | 4171 | RNAseq | 2.0246 | 0.0000 | |
| SRP133303 | MCM2 | 4171 | RNAseq | 1.9539 | 0.0000 | |
| SRP159526 | MCM2 | 4171 | RNAseq | 2.1075 | 0.0000 | |
| SRP193095 | MCM2 | 4171 | RNAseq | 1.7783 | 0.0000 | |
| SRP219564 | MCM2 | 4171 | RNAseq | 2.2861 | 0.0000 | |
| TCGA | MCM2 | 4171 | RNAseq | 0.9063 | 0.0000 |
Upregulated datasets: 17; Downregulated datasets: 0.
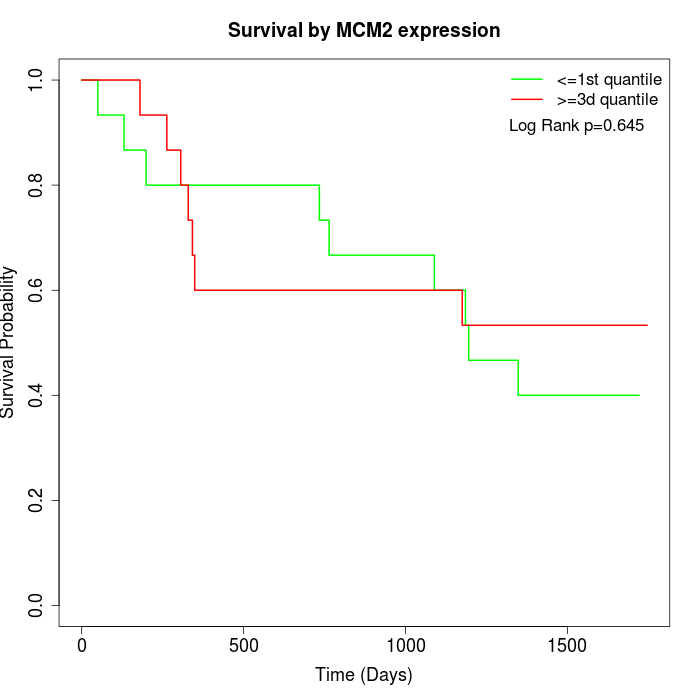
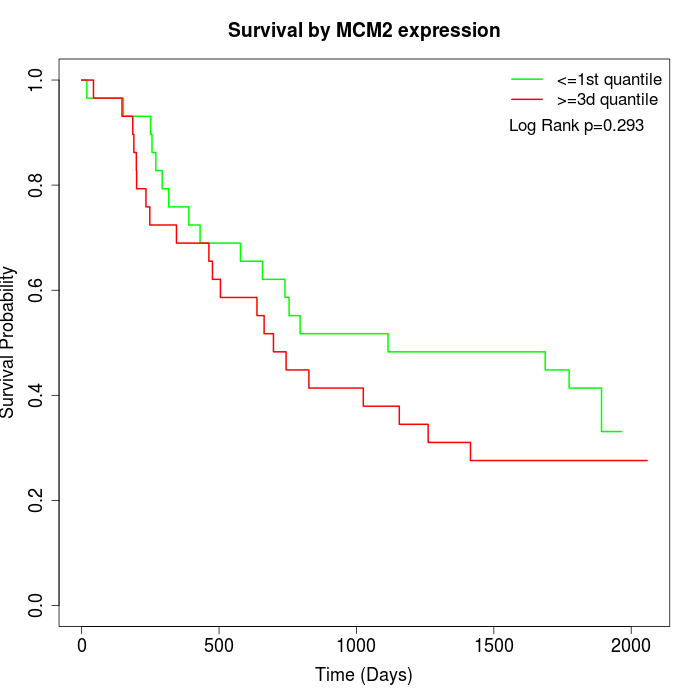
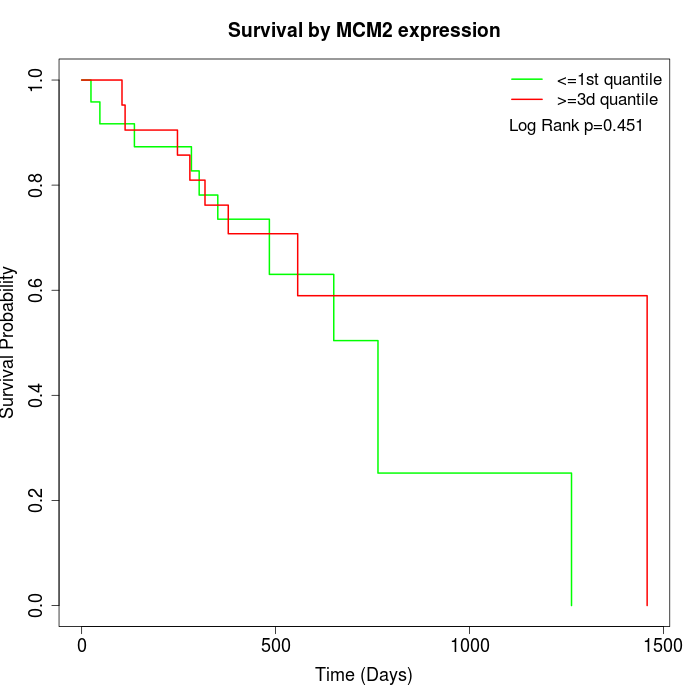
Survival by MCM2 expression:
Note: Click image to view full size file.
Copy number change of MCM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MCM2 | 4171 | 17 | 0 | 13 | |
| GSE20123 | MCM2 | 4171 | 17 | 0 | 13 | |
| GSE43470 | MCM2 | 4171 | 19 | 0 | 24 | |
| GSE46452 | MCM2 | 4171 | 14 | 5 | 40 | |
| GSE47630 | MCM2 | 4171 | 17 | 4 | 19 | |
| GSE54993 | MCM2 | 4171 | 2 | 7 | 61 | |
| GSE54994 | MCM2 | 4171 | 31 | 3 | 19 | |
| GSE60625 | MCM2 | 4171 | 0 | 6 | 5 | |
| GSE74703 | MCM2 | 4171 | 17 | 0 | 19 | |
| GSE74704 | MCM2 | 4171 | 13 | 0 | 7 | |
| TCGA | MCM2 | 4171 | 56 | 6 | 34 |
Total number of gains: 203; Total number of losses: 31; Total Number of normals: 254.
Somatic mutations of MCM2:
Generating mutation plots.
Highly correlated genes for MCM2:
Showing top 20/1381 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MCM2 | RFC4 | 0.868435 | 13 | 0 | 13 |
| MCM2 | MCM5 | 0.851208 | 12 | 0 | 12 |
| MCM2 | MCM6 | 0.847768 | 12 | 0 | 12 |
| MCM2 | UBE2C | 0.845594 | 12 | 0 | 12 |
| MCM2 | GINS2 | 0.845547 | 13 | 0 | 13 |
| MCM2 | RAD51AP1 | 0.841935 | 13 | 0 | 13 |
| MCM2 | NCAPH | 0.839345 | 9 | 0 | 9 |
| MCM2 | TIMELESS | 0.839165 | 12 | 0 | 12 |
| MCM2 | MCM7 | 0.837065 | 13 | 0 | 12 |
| MCM2 | DTL | 0.832998 | 12 | 0 | 12 |
| MCM2 | FOXM1 | 0.832819 | 13 | 0 | 12 |
| MCM2 | CDC45 | 0.832385 | 12 | 0 | 12 |
| MCM2 | MYBL2 | 0.832303 | 12 | 0 | 12 |
| MCM2 | GINS1 | 0.831213 | 11 | 0 | 11 |
| MCM2 | CCNB2 | 0.828543 | 11 | 0 | 11 |
| MCM2 | KIF4A | 0.823273 | 13 | 0 | 13 |
| MCM2 | CDCA3 | 0.822194 | 12 | 0 | 12 |
| MCM2 | BUB1B | 0.82114 | 11 | 0 | 11 |
| MCM2 | NEK2 | 0.81934 | 12 | 0 | 12 |
| MCM2 | TK1 | 0.816115 | 13 | 0 | 13 |
For details and further investigation, click here